A quantitative assessment of the dynamic modification of lipid-DNA probes on live cell membranes
- PMID: 32055389
- PMCID: PMC7003967
- DOI: 10.1039/c9sc04251b
A quantitative assessment of the dynamic modification of lipid-DNA probes on live cell membranes
Abstract
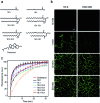
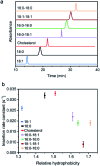
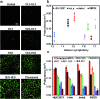
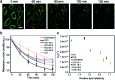
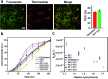
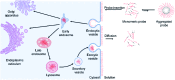
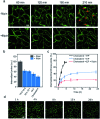
Synthetic lipid-DNA probes have recently attracted much attention for cell membrane analysis, transmembrane signal transduction, and regulating intercellular networks. These lipid-DNA probes can spontaneously insert onto plasma membranes simply after incubation. The highly precise and controllable DNA interactions have further allowed the programmable manipulation of these membrane-anchored functional probes. However, we still have quite limited understanding of how these lipid-DNA probes interact with cell membranes and also what parameters determine this process. In this study, we have systematically studied the dynamic process of cell membrane modification with a group of lipid-DNA probes. Our results indicated that the hydrophobicity of the lipid-DNA probes is strongly correlated with their membrane insertion and departure rates. Most cell membrane insertion stems from the monomeric form of probes, rather than the aggregates. Lipid-DNA probes can be removed from cell membranes through either endocytosis or direct outflow into the solution. As a result, long-term probe modifications on cell membranes can be realized in the presence of excess probes in the solution and/or endocytosis inhibitors. For the first time, we have successfully improved the membrane persistence of lipid-DNA probes to more than 24 h. Our quantitative data have dramatically improved our understanding of how lipid-DNA probes dynamically interact with cell membranes. These results can be further used to allow a broad range of applications of lipid-DNA probes for cell membrane analysis and regulation.
This journal is © The Royal Society of Chemistry 2019.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

