Hepatocyte nuclear factor 4α negatively regulates connective tissue growth factor during liver regeneration
- PMID: 32057145
- PMCID: PMC7722640
- DOI: 10.1096/fj.201902382R
Hepatocyte nuclear factor 4α negatively regulates connective tissue growth factor during liver regeneration
Abstract
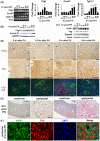
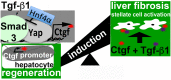
Liver regeneration after injury requires fine-tune regulation of connective tissue growth factor (Ctgf). It also involves dynamic expression of hepatocyte nuclear factor (Hnf)4α, Yes-associated protein (Yap), and transforming growth factor (Tgf)-β. The upstream inducers of Ctgf, such as Yap, etc, are well-known. However, the negative regulator of Ctgf remains unclear. Here, we investigated the Hnf4α regulation of Ctgf post-various types of liver injury. Both wild-type animals and animals contained siRNA-mediated Hnf4α knockdown and Cre-mediated Ctgf conditional deletion were used. We observed that Ctgf induction was associated with Hnf4α decline, nuclear Yap accumulation, and Tgf-β upregulation during early stage of liver regeneration. The Ctgf promoter contained an Hnf4α binding sequence that overlapped with the cis-regulatory element for Yap and Tgf-β. Ctgf loss attenuated inflammation, hepatocyte proliferation, and collagen synthesis, whereas Hnf4α knockdown enhanced Ctgf induction and liver fibrogenesis. These findings provided a new mechanism about fine-tuned regulation of Ctgf through Hnf4α antagonism of Yap and Tgf-β activities to balance regenerative and fibrotic signals.
Keywords: connective tissue growth factor (Ctgf); hepatocyte nuclear factor 4α (Hnf4α); liver injury; liver regeneration.
© 2020 Federation of American Societies for Experimental Biology.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding publication of this manuscript.
Figures




Similar articles
-
S1P Stimulates Proliferation by Upregulating CTGF Expression through S1PR2-Mediated YAP Activation.Mol Cancer Res. 2018 Oct;16(10):1543-1555. doi: 10.1158/1541-7786.MCR-17-0681. Epub 2018 Jun 14. Mol Cancer Res. 2018. PMID: 29903770
-
YAP integrates the regulatory Snail/HNF4α circuitry controlling epithelial/hepatocyte differentiation.Cell Death Dis. 2019 Oct 10;10(10):768. doi: 10.1038/s41419-019-2000-8. Cell Death Dis. 2019. PMID: 31601778 Free PMC article.
-
Taurocholate Induces Connective Tissue Growth Factor Expression in Hepatocytes Through ERK-YAP Signaling.Cell Physiol Biochem. 2018;50(5):1711-1725. doi: 10.1159/000494790. Epub 2018 Nov 1. Cell Physiol Biochem. 2018. PMID: 30384360
-
Role of hepatocyte nuclear factor 4α (HNF4α) in cell proliferation and cancer.Gene Expr. 2015;16(3):101-8. doi: 10.3727/105221615X14181438356292. Gene Expr. 2015. PMID: 25700366 Free PMC article. Review.
-
Connective tissue growth factor: a fibrogenic master switch in fibrotic liver diseases.Liver Int. 2008 Sep;28(8):1065-79. doi: 10.1111/j.1478-3231.2008.01826.x. Liver Int. 2008. PMID: 18783549 Review.
Cited by
-
Small Molecule-Induced Differentiation As a Potential Therapy for Liver Cancer.Adv Sci (Weinh). 2022 May;9(15):e2103619. doi: 10.1002/advs.202103619. Epub 2022 Mar 27. Adv Sci (Weinh). 2022. PMID: 35343115 Free PMC article.
-
Molecular pathways of liver regeneration: A comprehensive review.World J Hepatol. 2021 Mar 27;13(3):270-290. doi: 10.4254/wjh.v13.i3.270. World J Hepatol. 2021. PMID: 33815672 Free PMC article. Review.
-
CCN2/CTGF promotes liver fibrosis through crosstalk with the Slit2/Robo signaling.J Cell Commun Signal. 2023 Mar;17(1):137-150. doi: 10.1007/s12079-022-00713-y. Epub 2022 Dec 5. J Cell Commun Signal. 2023. PMID: 36469291 Free PMC article.
-
Methyl 6-O-cinnamoyl-α-d-glucopyranoside Ameliorates Acute Liver Injury by Inhibiting Oxidative Stress Through the Activation of Nrf2 Signaling Pathway.Front Pharmacol. 2022 Apr 26;13:873938. doi: 10.3389/fphar.2022.873938. eCollection 2022. Front Pharmacol. 2022. PMID: 35559264 Free PMC article.
-
Pseudogene RPL32P3 regulates the blood-tumor barrier permeability via the YBX2/HNF4G axis.Cell Death Discov. 2021 Nov 24;7(1):367. doi: 10.1038/s41420-021-00758-9. Cell Death Discov. 2021. PMID: 34819492 Free PMC article.
References
-
- Nwidu LL, Oboma YI. Telfairia occidentalis (Cucurbitaceae) pulp extract mitigates rifampicin‐isoniazid‐induced hepatotoxicity in an in vivo rat model of oxidative stress. J Integr Med. 2019;17:46‐56. - PubMed
-
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. J Hepatol. 2012;57:692‐694. - PubMed
-
- Ling CQ, Fan J, Lin HS, et al. Clinical practice guidelines for the treatment of primary liver cancer with integrative traditional Chinese and Western medicine. J Integr Med. 2018;16:236‐248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

