Targeting glioma-initiating cells via the tyrosine metabolic pathway
- PMID: 32059178
- PMCID: PMC8447888
- DOI: 10.3171/2019.11.JNS192028
Targeting glioma-initiating cells via the tyrosine metabolic pathway
Abstract
Objective: Despite an aggressive multimodal therapeutic regimen, glioblastoma (GBM) continues to portend a grave prognosis, which is driven in part by tumor heterogeneity at both the molecular and cellular levels. Accordingly, herein the authors sought to identify metabolic differences between GBM tumor core cells and edge cells and, in so doing, elucidate novel actionable therapeutic targets centered on tumor metabolism.
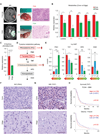
Methods: Comprehensive metabolic analyses were performed on 20 high-grade glioma (HGG) tissues and 30 glioma-initiating cell (GIC) sphere culture models. The results of the metabolic analyses were combined with the Ivy GBM data set. Differences in tumor metabolism between GBM tumor tissue derived from within the contrast-enhancing region (i.e., tumor core) and that from the peritumoral brain lesions (i.e., tumor edge) were sought and explored. Such changes were ultimately confirmed at the protein level via immunohistochemistry.
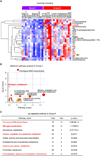
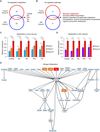
Results: Metabolic heterogeneity in both HGG tumor tissues and GBM sphere culture models was identified, and analyses suggested that tyrosine metabolism may serve as a possible therapeutic target in GBM, particularly in the tumor core. Furthermore, activation of the enzyme tyrosine aminotransferase (TAT) within the tyrosine metabolic pathway influenced the noted therapeutic resistance of the GBM core.
Conclusions: Selective inhibition of the tyrosine metabolism pathway may prove highly beneficial as an adjuvant to multimodal GBM therapies.
Keywords: glioblastoma; heterogeneity; high-grade glioma; metabolism; nitrogen metabolism; oncology; tyrosine aminotransferase.
Figures



References
-
- Ahluwalia MS, Patton C, Stevens G, Tekautz T, Angelov L, Vogelbaum MA, et al.: Phase II trial of ritonavir/lopinavir in patients with progressive or recurrent high-grade gliomas. J Neurooncol 102:317–321, 2011 - PubMed
-
- Aisenberg AC, Reinafarje B, Potter VR: Studies on the Pasteur effect. I. General observations. J Biol Chem 224:1099–1113, 1957 - PubMed
-
- Argilés JM, Costelli P, Carbó N, Pallarés-Trujillo J, López-Soriano FJ: Tumour growth and nitrogen metabolism in the host. Int J Oncol 14:479–486, 1999 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

