The modular nature of protein evolution: domain rearrangement rates across eukaryotic life
- PMID: 32059645
- PMCID: PMC7023805
- DOI: 10.1186/s12862-020-1591-0
The modular nature of protein evolution: domain rearrangement rates across eukaryotic life
Abstract
Background: Modularity is important for evolutionary innovation. The recombination of existing units to form larger complexes with new functionalities spares the need to create novel elements from scratch. In proteins, this principle can be observed at the level of protein domains, functional subunits which are regularly rearranged to acquire new functions.
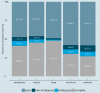
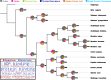
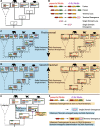
Results: In this study we analyse the mechanisms leading to new domain arrangements in five major eukaryotic clades (vertebrates, insects, fungi, monocots and eudicots) at unprecedented depth and breadth. This allows, for the first time, to directly compare rates of rearrangements between different clades and identify both lineage specific and general patterns of evolution in the context of domain rearrangements. We analyse arrangement changes along phylogenetic trees by reconstructing ancestral domain content in combination with feasible single step events, such as fusion or fission. Using this approach we explain up to 70% of all rearrangements by tracing them back to their precursors. We find that rates in general and the ratio between these rates for a given clade in particular, are highly consistent across all clades. In agreement with previous studies, fusions are the most frequent event leading to new domain arrangements. A lineage specific pattern in fungi reveals exceptionally high loss rates compared to other clades, supporting recent studies highlighting the importance of loss for evolutionary innovation. Furthermore, our methodology allows us to link domain emergences at specific nodes in the phylogenetic tree to important functional developments, such as the origin of hair in mammals.
Conclusions: Our results demonstrate that domain rearrangements are based on a canonical set of mutational events with rates which lie within a relatively narrow and consistent range. In addition, gained knowledge about these rates provides a basis for advanced domain-based methodologies for phylogenetics and homology analysis which complement current sequence-based methods.
Keywords: Ancestral reconstruction; Evolutionary history; Protein domain; Proteome analysis; Rearrangement rates.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):427–32. doi: 10.1093/nar/gky995. - DOI - PMC - PubMed
-
- Forslund K, Sonnhammer ELL. Evolution of protein domain architectures. In: Anisimova M, editor. Evolutionary Genomics: Statistical and Computational Methods, Volume 2. Totowa, NJ: Humana Press; 2012.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

