Click Chemistry in Proteomic Investigations
- PMID: 32059777
- PMCID: PMC7087397
- DOI: 10.1016/j.cell.2020.01.025
Click Chemistry in Proteomic Investigations
Abstract
Despite advances in genetic and proteomic techniques, a complete portrait of the proteome and its complement of dynamic interactions and modifications remains a lofty, and as of yet, unrealized, objective. Specifically, traditional biological and analytical approaches have not been able to address key questions relating to the interactions of proteins with small molecules, including drugs, drug candidates, metabolites, or protein post-translational modifications (PTMs). Fortunately, chemists have bridged this experimental gap through the creation of bioorthogonal reactions. These reactions allow for the incorporation of chemical groups with highly selective reactivity into small molecules or protein modifications without perturbing their biological function, enabling the selective installation of an analysis tag for downstream investigations. The introduction of chemical strategies to parse and enrich subsets of the "functional" proteome has empowered mass spectrometry (MS)-based methods to delve more deeply and precisely into the biochemical state of cells and its perturbations by small molecules. In this Primer, we discuss how one of the most versatile bioorthogonal reactions, "click chemistry", has been exploited to overcome limitations of biological approaches to enable the selective marking and functional investigation of critical protein-small-molecule interactions and PTMs in native biological environments.
Copyright © 2020 Elsevier Inc. All rights reserved.
Figures



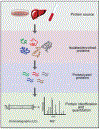


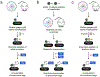

References
-
- Abegg D, Frei R, Cerato L, Hari DP, Wang C, Waser J, and Adibekian A (2015). Proteome-wide profiling of targets of cysteine reactive small molecules by using ethynyl benziodoxolone reagents. Angewandte Chemie-International Edition 54, 10852–10857. - PubMed
-
- Abo M, and Weerapana E (2015). A caged electrophilic probe for global analysis of cysteine reactivity in living cells. J Am Chem Soc 137, 7087–7090. - PubMed
-
- Aebi M (2013). N-linked protein glycosylation in the er. Biochimica et biophysica acta 1833, 2430–2437. - PubMed
-
- Agard NJ, Prescher JA, and Bertozzi CR (2004). A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc 126, 15046–15047. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

