Dissimilarity of the gut-lung axis and dysbiosis of the lower airways in ventilated preterm infants
- PMID: 32060060
- PMCID: PMC7236867
- DOI: 10.1183/13993003.01909-2019
Dissimilarity of the gut-lung axis and dysbiosis of the lower airways in ventilated preterm infants
Abstract
Background: Chronic lung disease of prematurity (CLD), also called bronchopulmonary dysplasia, is a major consequence of preterm birth, but the role of the microbiome in its development remains unclear. Therefore, we assessed the progression of the bacterial community in ventilated preterm infants over time in the upper and lower airways, and assessed the gut-lung axis by comparing bacterial communities in the upper and lower airways with stool findings. Finally, we assessed whether the bacterial communities were associated with lung inflammation to suggest dysbiosis.
Methods: We serially sampled multiple anatomical sites including the upper airway (nasopharyngeal aspirates), lower airways (tracheal aspirate fluid and bronchoalveolar lavage fluid) and the gut (stool) of ventilated preterm-born infants. Bacterial DNA load was measured in all samples and sequenced using the V3-V4 region of the 16S rRNA gene.
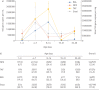
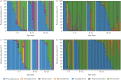
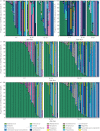
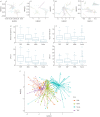
Results: From 1102 (539 nasopharyngeal aspirates, 276 tracheal aspirate fluid, 89 bronchoalveolar lavage, 198 stool) samples from 55 preterm infants, 352 (32%) amplified suitably for 16S RNA gene sequencing. Bacterial load was low at birth and quickly increased with time, but was associated with predominant operational taxonomic units (OTUs) in all sample types. There was dissimilarity in bacterial communities between the upper and lower airways and the gut, with a separate dysbiotic inflammatory process occurring in the lower airways of infants. Individual OTUs were associated with increased inflammatory markers.
Conclusions: Taken together, these findings suggest that targeted treatment of the predominant organisms, including those not routinely treated, such as Ureaplasma spp., may decrease the development of CLD in preterm-born infants.
Copyright ©ERS 2020.
Conflict of interest statement
Conflict of interest: D. Gallacher has nothing to disclose. Conflict of interest: E. Mitchell has nothing to disclose. Conflict of interest: D. Alber has nothing to disclose. Conflict of interest: R. Wach has nothing to disclose. Conflict of interest: N. Klein has nothing to disclose. Conflict of interest: J.R. Marchesi has nothing to disclose. Conflict of interest: S. Kotecha has nothing to disclose.
Figures

Comment in
-
Bronchopulmonary dysplasia: a crime of opportunity?Eur Respir J. 2020 May 7;55(5):2000551. doi: 10.1183/13993003.00551-2020. Print 2020 May. Eur Respir J. 2020. PMID: 32381633 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
