On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability
- PMID: 32060097
- PMCID: PMC7105313
- DOI: 10.1074/jbc.REV119.007678
On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability
Abstract
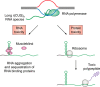
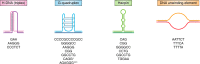
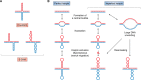
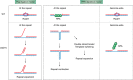
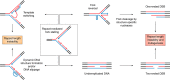
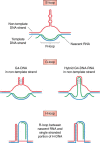
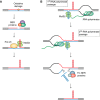
Expansions of simple tandem repeats are responsible for almost 50 human diseases, the majority of which are severe, degenerative, and not currently treatable or preventable. In this review, we first describe the molecular mechanisms of repeat-induced toxicity, which is the connecting link between repeat expansions and pathology. We then survey alternative DNA structures that are formed by expandable repeats and review the evidence that formation of these structures is at the core of repeat instability. Next, we describe the consequences of the presence of long structure-forming repeats at the molecular level: somatic and intergenerational instability, fragility, and repeat-induced mutagenesis. We discuss the reasons for gender bias in intergenerational repeat instability and the tissue specificity of somatic repeat instability. We also review the known pathways in which DNA replication, transcription, DNA repair, and chromatin state interact and thereby promote repeat instability. We then discuss possible reasons for the persistence of disease-causing DNA repeats in the genome. We describe evidence suggesting that these repeats are a payoff for the advantages of having abundant simple-sequence repeats for eukaryotic genome function and evolvability. Finally, we discuss two unresolved fundamental questions: (i) why does repeat behavior differ between model systems and human pedigrees, and (ii) can we use current knowledge on repeat instability mechanisms to cure repeat expansion diseases?
Keywords: DNA recombination; DNA repair; DNA replication; DNA structure; G-quadruplex; Huntington disease; R-loop; S-DNA; amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease); gene expression; genomic instability; hairpin; trinucleotide repeat disease; triplex H-DNA.
© 2020 Khristich and Mirkin.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
The balancing act of DNA repeat expansions.Curr Opin Genet Dev. 2013 Jun;23(3):280-8. doi: 10.1016/j.gde.2013.04.009. Epub 2013 May 29. Curr Opin Genet Dev. 2013. PMID: 23725800 Free PMC article. Review.
-
Repeat instability during DNA repair: Insights from model systems.Crit Rev Biochem Mol Biol. 2015 Mar-Apr;50(2):142-67. doi: 10.3109/10409238.2014.999192. Epub 2015 Jan 22. Crit Rev Biochem Mol Biol. 2015. PMID: 25608779 Free PMC article. Review.
-
R-loops: targets for nuclease cleavage and repeat instability.Curr Genet. 2018 Aug;64(4):789-794. doi: 10.1007/s00294-018-0806-z. Epub 2018 Jan 11. Curr Genet. 2018. PMID: 29327083 Free PMC article. Review.
-
DNA structures, repeat expansions and human hereditary disorders.Curr Opin Struct Biol. 2006 Jun;16(3):351-8. doi: 10.1016/j.sbi.2006.05.004. Epub 2006 May 19. Curr Opin Struct Biol. 2006. PMID: 16713248 Review.
-
Role of recombination and replication fork restart in repeat instability.DNA Repair (Amst). 2017 Aug;56:156-165. doi: 10.1016/j.dnarep.2017.06.018. Epub 2017 Jun 9. DNA Repair (Amst). 2017. PMID: 28641941 Free PMC article. Review.
Cited by
-
Efficacy Evaluation of Treatment of Psoriasis Via Narrow Band-Ultraviolet Radiation.J Lasers Med Sci. 2024 Jul 27;15:e26. doi: 10.34172/jlms.2024.26. eCollection 2024. J Lasers Med Sci. 2024. PMID: 39188934 Free PMC article.
-
Heterogeneous migration routes of DNA triplet repeat slip-outs.Biophys Rep (N Y). 2022 Sep 14;2(3):None. doi: 10.1016/j.bpr.2022.100070. Biophys Rep (N Y). 2022. PMID: 36299495 Free PMC article.
-
Stable G-quadruplex DNA structures promote replication-dependent genome instability.J Biol Chem. 2022 Jun;298(6):101947. doi: 10.1016/j.jbc.2022.101947. Epub 2022 Apr 18. J Biol Chem. 2022. PMID: 35447109 Free PMC article.
-
Contractions of the C-Terminal Domain of Saccharomyces cerevisiae Rpb1p Are Mediated by Rad5p.G3 (Bethesda). 2020 Jul 7;10(7):2543-2551. doi: 10.1534/g3.120.401409. G3 (Bethesda). 2020. PMID: 32467128 Free PMC article.
-
Rad9-mediated checkpoint activation is responsible for elevated expansions of GAA repeats in CST-deficient yeast.Genetics. 2021 Oct 2;219(2):iyab125. doi: 10.1093/genetics/iyab125. Genetics. 2021. PMID: 34849883 Free PMC article.
References
-
- Nettleship E. (1905) On heredity in the various forms of cataract. Rep. R. Lond. Ophthal. Hosp. 16, 179–246
-
- Fleischer B. (1918) Über myotonische Dystrophie mit Katarakt. Graefes. Arch. Klein. Ophthalmol. 96, 91–133 10.1007/BF02018704 - DOI
-
- Sherman S. L., Jacobs P. A., Morton N. E., Froster-Iskenius U., Howard-Peebles P. N., Nielsen K. B., Partington M. W., Sutherland G. R., Turner G., and Watson M. (1985) Further segregation analysis of the fragile X syndrome with special reference to transmitting males. Hum. Genet. 69, 289–299 10.1007/BF00291644 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

