Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma
- PMID: 32060274
- PMCID: PMC7021791
- DOI: 10.1038/s41467-020-14639-9
Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma
Abstract
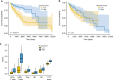
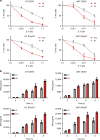
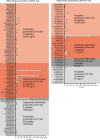
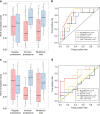
Predicting the outcome of immunotherapy treatment in melanoma patients is challenging. Alterations in genes involved in antigen presentation and the interferon gamma (IFNγ) pathway play an important role in the immune response to tumors. We describe here that the overexpression of PSMB8 and PSMB9, two major components of the immunoproteasome, is predictive of better survival and improved response to immune-checkpoint inhibitors of melanoma patients. We study the mechanism underlying this connection by analyzing the antigenic peptide repertoire of cells that overexpress these subunits using HLA peptidomics. We find a higher response of patient-matched tumor infiltrating lymphocytes against antigens diferentially presented after immunoproteasome overexpression. Importantly, we find that PSMB8 and PSMB9 expression levels are much stronger predictors of melanoma patients' immune response to checkpoint inhibitors than the tumors' mutational burden. These results suggest that PSMB8 and PSMB9 expression levels can serve as important biomarkers for stratifying melanoma patients for immune-checkpoint treatment.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

