Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff
- PMID: 32060277
- PMCID: PMC7021729
- DOI: 10.1038/s41467-020-14724-z
Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff
Abstract
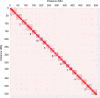
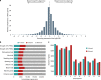
Teff (Eragrostis tef) is a cornerstone of food security in the Horn of Africa, where it is prized for stress resilience, grain nutrition, and market value. Here, we report a chromosome-scale assembly of allotetraploid teff (variety Dabbi) and patterns of subgenome dynamics. The teff genome contains two complete sets of homoeologous chromosomes, with most genes maintaining as syntenic gene pairs. TE analysis allows us to estimate that the teff polyploidy event occurred ~1.1 million years ago (mya) and that the two subgenomes diverged ~5.0 mya. Despite this divergence, we detect no large-scale structural rearrangements, homoeologous exchanges, or biased gene loss, in contrast to many other allopolyploids. The two teff subgenomes have partitioned their ancestral functions based on divergent expression across a diverse expression atlas. Together, these genomic resources will be useful for accelerating breeding of this underutilized grain crop and for fundamental insights into polyploid genome evolution.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- CSA. Agricultural Sample Survey 2011/2012: Report on Area and Production of Major Crops (Central Statistical Agency Addis Ababa, 2012).
-
- Stallknecht, G. F., Gilbertson, K. M. & Eckhoff, J. in New crops 231–234 (Wiley, New York, 1993).
-
- Demissie, A. in Narrowing the Rift. Tef Research and Development. Proc. International Workshop on Tef Genetics and Improvement (Debre Zeit, Ethiopia, 2000).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

