A slipped-CAG DNA-binding small molecule induces trinucleotide-repeat contractions in vivo
- PMID: 32060489
- PMCID: PMC7043212
- DOI: 10.1038/s41588-019-0575-8
A slipped-CAG DNA-binding small molecule induces trinucleotide-repeat contractions in vivo
Abstract
In many repeat diseases, such as Huntington's disease (HD), ongoing repeat expansions in affected tissues contribute to disease onset, progression and severity. Inducing contractions of expanded repeats by exogenous agents is not yet possible. Traditional approaches would target proteins driving repeat mutations. Here we report a compound, naphthyridine-azaquinolone (NA), that specifically binds slipped-CAG DNA intermediates of expansion mutations, a previously unsuspected target. NA efficiently induces repeat contractions in HD patient cells as well as en masse contractions in medium spiny neurons of HD mouse striatum. Contractions are specific for the expanded allele, independently of DNA replication, require transcription across the coding CTG strand and arise by blocking repair of CAG slip-outs. NA-induced contractions depend on active expansions driven by MutSβ. NA injections in HD mouse striatum reduce mutant HTT protein aggregates, a biomarker of HD pathogenesis and severity. Repeat-structure-specific DNA ligands are a novel avenue to contract expanded repeats.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Figures



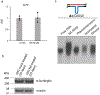


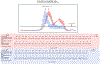
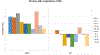









Comment in
-
A small molecule kicks repeat expansion into reverse.Nat Genet. 2020 Feb;52(2):136-137. doi: 10.1038/s41588-020-0577-6. Nat Genet. 2020. PMID: 32060488 No abstract available.
References
-
- Kennedy L et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet 12, 3359–67 (2003). - PubMed
-
- Lopez Castel A, Cleary JD & Pearson CE Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol 11, 165–70 (2010). - PubMed
-
- Mirkin SM Expandable DNA repeats and human disease. Nature 447, 932–40 (2007). - PubMed
-
- Pearson CE, Nichol Edamura K & Cleary JD Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6, 729–42 (2005). - PubMed
-
- Sathasivam K, Amaechi I, Mangiarini L & Bates G Identification of an HD patient with a (CAG)180 repeat expansion and the propagation of highly expanded CAG repeats in lambda phage. Hum Genet 99, 692–5 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

