Characterization of Drosophila octopamine receptor neuronal expression using MiMIC-converted Gal4 lines
- PMID: 32060912
- PMCID: PMC7998515
- DOI: 10.1002/cne.24883
Characterization of Drosophila octopamine receptor neuronal expression using MiMIC-converted Gal4 lines
Abstract
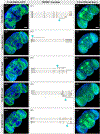
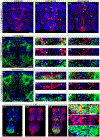
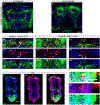
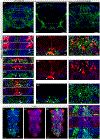
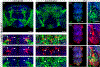
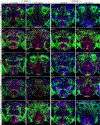
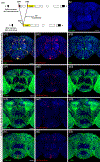
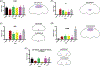
Octopamine, the invertebrate analog of norepinephrine, is known to modulate a large variety of behaviors in Drosophila including feeding initiation, locomotion, aggression, and courtship, among many others. Significantly less is known about the identity of the neurons that receive octopamine input and how they mediate octopamine-regulated behaviors. Here, we characterize adult neuronal expression of MiMIC-converted Trojan-Gal4 lines for each of the five Drosophila octopamine receptors. Broad neuronal expression was observed for all five octopamine receptors, yet distinct differences among them were also apparent. Use of immunostaining for the octopamine neurotransmitter synthesis enzyme Tdc2, along with a novel genome-edited conditional Tdc2-LexA driver, revealed all five octopamine receptors express in Tdc2/octopamine neurons to varying degrees. This suggests autoreception may be an important circuit mechanism by which octopamine modulates behavior.
Keywords: Drosophila; RRID:AB_221568; RRID:AB_2340686; RRID:AB_2340850; RRID:AB_2536611; RRID:AB_2633280; RRID:AB_2814891; RRID:BDSC_27392; RRID:BDSC_42119; RRID:BDSC_43050; RRID:BDSC_57940; RRID:BDSC_59133; RRID:BDSC_60312; RRID:BDSC_60313; RRID:BDSC_67636; RRID:BDSC_68264; autoreception; octopamine; octopamine receptor.
© 2020 Wiley Periodicals, Inc.
Figures



Similar articles
-
A map of sensilla and neurons in the taste system of drosophila larvae.J Comp Neurol. 2017 Dec 15;525(18):3865-3889. doi: 10.1002/cne.24308. Epub 2017 Sep 11. J Comp Neurol. 2017. PMID: 28842919
-
Development of the anterior visual input pathway to the Drosophila central complex.J Comp Neurol. 2017 Nov 1;525(16):3458-3475. doi: 10.1002/cne.24277. Epub 2017 Aug 21. J Comp Neurol. 2017. PMID: 28675433 Free PMC article.
-
Drosophila ammonium transporter Rh50 is required for integrity of larval muscles and neuromuscular system.J Comp Neurol. 2020 Jan 1;528(1):81-94. doi: 10.1002/cne.24742. Epub 2019 Jul 18. J Comp Neurol. 2020. PMID: 31273786
-
Structure and development of the subesophageal zone of the Drosophila brain. II. Sensory compartments.J Comp Neurol. 2018 Jan 1;526(1):33-58. doi: 10.1002/cne.24316. Epub 2017 Sep 28. J Comp Neurol. 2018. PMID: 28875566 Free PMC article.
-
Insect octopamine receptors: a new classification scheme based on studies of cloned Drosophila G-protein coupled receptors.Invert Neurosci. 2005 Nov;5(3-4):111-8. doi: 10.1007/s10158-005-0001-z. Epub 2005 Oct 24. Invert Neurosci. 2005. PMID: 16211376 Review.
Cited by
-
Two classes of amine/glutamate multi-transmitter neurons innervate Drosophila internal male reproductive organs.bioRxiv [Preprint]. 2025 Jul 28:2025.07.23.666348. doi: 10.1101/2025.07.23.666348. bioRxiv. 2025. PMID: 40766443 Free PMC article. Preprint.
-
Monoamine-induced diacylglycerol signaling rapidly accumulates Unc13 in nanoclusters for fast presynaptic potentiation.Proc Natl Acad Sci U S A. 2025 Aug 26;122(34):e2514151122. doi: 10.1073/pnas.2514151122. Epub 2025 Aug 20. Proc Natl Acad Sci U S A. 2025. PMID: 40833403 Free PMC article.
-
Expression and potential regulatory functions of Drosophila octopamine receptors in the female reproductive tract.G3 (Bethesda). 2024 Mar 6;14(3):jkae012. doi: 10.1093/g3journal/jkae012. G3 (Bethesda). 2024. PMID: 38244217 Free PMC article.
-
Regulation of Drosophila oviduct muscle contractility by octopamine.iScience. 2022 Jul 2;25(8):104697. doi: 10.1016/j.isci.2022.104697. eCollection 2022 Aug 19. iScience. 2022. PMID: 35880044 Free PMC article.
-
Constitutive and Conditional Epitope Tagging of Endogenous G-Protein-Coupled Receptors in Drosophila.J Neurosci. 2024 Aug 14;44(33):e2377232024. doi: 10.1523/JNEUROSCI.2377-23.2024. J Neurosci. 2024. PMID: 38937100 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

