Capsule Networks Showed Excellent Performance in the Classification of hERG Blockers/Nonblockers
- PMID: 32063849
- PMCID: PMC6997788
- DOI: 10.3389/fphar.2019.01631
Capsule Networks Showed Excellent Performance in the Classification of hERG Blockers/Nonblockers
Abstract
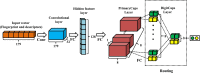
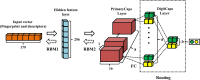
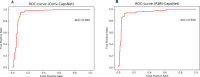
Capsule networks (CapsNets), a new class of deep neural network architectures proposed recently by Hinton et al., have shown a great performance in many fields, particularly in image recognition and natural language processing. However, CapsNets have not yet been applied to drug discovery-related studies. As the first attempt, we in this investigation adopted CapsNets to develop classification models of hERG blockers/nonblockers; drugs with hERG blockade activity are thought to have a potential risk of cardiotoxicity. Two capsule network architectures were established: convolution-capsule network (Conv-CapsNet) and restricted Boltzmann machine-capsule networks (RBM-CapsNet), in which convolution and a restricted Boltzmann machine (RBM) were used as feature extractors, respectively. Two prediction models of hERG blockers/nonblockers were then developed by Conv-CapsNet and RBM-CapsNet with the Doddareddy's training set composed of 2,389 compounds. The established models showed excellent performance in an independent test set comprising 255 compounds, with prediction accuracies of 91.8 and 92.2% for Conv-CapsNet and RBM-CapsNet models, respectively. Various comparisons were also made between our models and those developed by other machine learning methods including deep belief network (DBN), convolutional neural network (CNN), multilayer perceptron (MLP), support vector machine (SVM), k-nearest neighbors (kNN), logistic regression (LR), and LightGBM, and with different training sets. All the results showed that the models by Conv-CapsNet and RBM-CapsNet are among the best classification models. Overall, the excellent performance of capsule networks achieved in this investigation highlights their potential in drug discovery-related studies.
Keywords: Capsule network; classification model; convolution-capsule network; deep learning; hERG; restricted Boltzmann machine-capsule networks.
Copyright © 2020 Wang, Huang, Jiang, Wang, Zou, Fu and Yang.
Figures
Comment in
-
Artificial intelligence (AI) is paving the way for a critical role in drug discovery, drug design, and studying drug-drug interactions - correspondence.Int J Surg. 2023 Oct 1;109(10):3242-3244. doi: 10.1097/JS9.0000000000000564. Int J Surg. 2023. PMID: 37352517 Free PMC article. No abstract available.
References
-
- Afshar P., Mohammadi A., Plataniotis K. N. (2018). “Brain tumor type classification via capsule networks,” in 2018 25th IEEE International Conference on Image Processing (ICIP) (Athens: IEEE; ), 3129–3133. 10.1109/icip.2018.8451379 - DOI
-
- Bengio Y. (2009). Learning deep architectures for AI. Found. Trends Mach. Learn. 2, 1–127. 10.1561/2200000006 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

