miR-193a-5p promotes pancreatic cancer cell metastasis through SRSF6-mediated alternative splicing of OGDHL and ECM1
- PMID: 32064152
- PMCID: PMC7017744
miR-193a-5p promotes pancreatic cancer cell metastasis through SRSF6-mediated alternative splicing of OGDHL and ECM1
Abstract
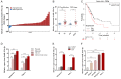
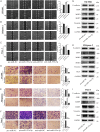
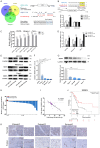
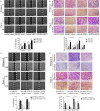
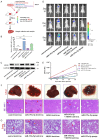
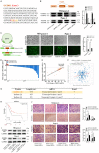
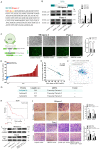
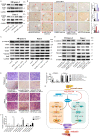
MicroRNAs (miRNAs) are short and non-coding RNAs binding to 3'UTR of target mRNAs to downregulate their expression. Recent studies have shown that miRNAs indirectly regulated alternative splicing (AS) by targeting splicing factors and caused shifts in splicing patterns of target genes. However, the roles of miRNA-regulating splicing factors in pancreatic cancer progression remain unknown. Herein, we reported that miR-193a-5p was markedly upregulated in pancreatic cancer tissues and cells and correlated with clinical outcomes of pancreatic cancer patients. Overexpression of miR-193a-5p contributed to the metastasis of pancreatic cancer cells both in vitro and in vivo. The mechanistic investigation suggested that miR-193a-5p modulated oxoglutarate dehydrogenase-like (OGDHL) and extracellular matrix protein 1 (ECM1) AS by targeting serine/arginine-rich splicing factor 6 (SRSF6), leading to the activation of the epithelial-to-mesenchymal transition (EMT) process. Together, our findings highlighted the role of miR-193a-5p-targeting SRSF6 in pancreatic cancer metastasis, which may serve as a novel target for pancreatic cancer diagnosis and therapy.
Keywords: ECM1; OGDHL; Pancreatic cancer; SRSF6; alternative splicing; invasion; miR-193a-5p; migration.
AJCR Copyright © 2020.
Conflict of interest statement
None.
Figures
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Konstantinidis IT, Warshaw AL, Allen JN, Blaszkowsky LS, Castillo CF, Deshpande V, Hong TS, Kwak EL, Lauwers GY, Ryan DP, Wargo JA, Lillemoe KD, Ferrone CR. Pancreatic ductal adenocarcinoma: is there a survival difference for R1 resections versus locally advanced unresectable tumors? What is a “true” R0 resection? Ann Surg. 2013;257:731–6. - PubMed
-
- Yoshida BA, Sokoloff MM, Welch DR, Rinker-Schaeffer CW. Metastasis-suppressor genes: a review and perspective on an emerging field. J Natl Cancer Inst. 2000;92:1717–30. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. - PubMed
-
- Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, Fitamant J, Jones PD, Ghanta KS, Kawano S, Nagle JM, Deshpande V, Boucher Y, Kato T, Chen JK, Willmann JK, Bardeesy N, Beachy PA. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111:E3091–100. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
