Wheat PP2C-a10 regulates seed germination and drought tolerance in transgenic Arabidopsis
- PMID: 32065246
- PMCID: PMC7165162
- DOI: 10.1007/s00299-020-02520-4
Wheat PP2C-a10 regulates seed germination and drought tolerance in transgenic Arabidopsis
Abstract
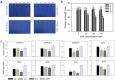
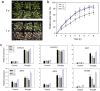
A wheat protein phosphatase PP2C-a10, which interacted with TaDOG1L1 and TaDOG1L4, promoted seed germination and decreased drought tolerance of transgenic Arabidopsis. Seed dormancy and germination are critical to plant fitness. DELAY OF GERMINATION 1 (DOG1) is a quantitative trait locus for dormancy in Arabidopsis thaliana. Some interactions between DOG1 and the type 2C protein phosphatases (PP2Cs) have been reported in Arabidopsis. However, the research on molecular functions and regulations of DOG1Ls and group A PP2Cs in wheat (Triticum aestivum. L), an important crop plant, is rare. In this study, the whole TaDOG1L family was identified. Expression analysis revealed that TaDOG1L2, TaDOG1L4 and TaDOG1L-N2 specially expressed in wheat grains, while others displayed distinct expression patterns. Yeast two-hybrid analysis of TaDOG1Ls and group A TaPP2Cs revealed interaction patterns differed from those in Arabidopsis, and TaDOG1L1 and TaDOG1L4 interacted with TaPP2C-a10. The qRT-PCR analysis showed that TaPP2C-a10 exhibited the highest transcript level in wheat grains. Further investigation showed that ectopic expression of TaPP2C-a10 in Arabidopsis promoted seed germination and decreased sensitivity to ABA during germination stage. Additionally, TaPP2C-a10 transgenic Arabidopsis exhibited decreased tolerance to drought stress. Finally, the phylogenetic analysis indicated that TaPP2C-a10 gene was conserved in angiosperm during evolutionary process. Overall, our results reveal the role of TaPP2C-a10 in seed germination and abiotic stress response, as well as the functional diversity of TaDOG1L family.
Keywords: ABA; DOG1; PP2C; Seed germination; Wheat.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures








References
-
- Ashikawa I, Abe F, Nakamura S. Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 2010;179:536–542. - PubMed
-
- Ashikawa I, Abe F, Nakamura S. DOG1-like genes in cereals: investigation of their function by means of ectopic expression in Arabidopsis. Plant Sci. 2013;208:1–9. - PubMed
-
- Ashikawa I, Mori M, Nakamura S, Abe F. A transgenic approach to controlling wheat seed dormancy level by using Triticeae DOG1-like genes. Transgenic Res. 2014;23:621–629. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

