Repression of viral gene expression and replication by the unfolded protein response effector XBP1u
- PMID: 32065579
- PMCID: PMC7082126
- DOI: 10.7554/eLife.51804
Repression of viral gene expression and replication by the unfolded protein response effector XBP1u
Abstract
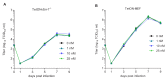
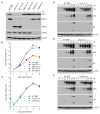
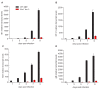
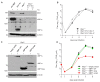
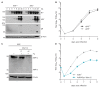
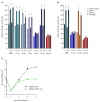
The unfolded protein response (UPR) is a cellular homeostatic circuit regulating protein synthesis and processing in the ER by three ER-to-nucleus signaling pathways. One pathway is triggered by the inositol-requiring enzyme 1 (IRE1), which splices the X-box binding protein 1 (Xbp1) mRNA, thereby enabling expression of XBP1s. Another UPR pathway activates the activating transcription factor 6 (ATF6). Here we show that murine cytomegalovirus (MCMV), a prototypic β-herpesvirus, harnesses the UPR to regulate its own life cycle. MCMV activates the IRE1-XBP1 pathway early post infection to relieve repression by XBP1u, the product of the unspliced Xbp1 mRNA. XBP1u inhibits viral gene expression and replication by blocking the activation of the viral major immediate-early promoter by XBP1s and ATF6. These findings reveal a redundant function of XBP1s and ATF6 as activators of the viral life cycle, and an unexpected role of XBP1u as a potent repressor of both XBP1s and ATF6-mediated activation.
Keywords: ATF6; XBP1u; cell biology; infectious disease; microbiology; mouse; murid herpesvirus 1; transcription factor; unfolded protein response; virus.
Plain language summary
Cells survive by making many different proteins that each carry out specific tasks. To work correctly, each protein must be made and then folded into the right shape. Cells carefully monitor protein folding because unfolded proteins can compromise their viability. A protein called XBP1 is important in controlling how cells respond to unfolded proteins. Normally, cells contain a form of this protein called XBP1u, while increasing numbers of unfolded proteins trigger production of a form called XBP1s. The change from one form to the other is activated by a protein called IRE1. Viruses often manipulate stress responses like the unfolded protein response to help take control of the cell and produce more copies of the virus. Murine cytomegalovirus, which is known as MCMV for short, is a herpes-like virus that infects mice; it stops IRE1 activation and XBP1s production during the later stages of infection. However, research had shown that the unfolded protein response was triggered for a short time at an early stage of infection with MCMV, and it was unclear why this might be. Hinte et al. studied the effect of MCMV on cells grown in the laboratory. The experiments showed that a small dose of cell stress, namely activating the unfolded protein response briefly during early infection, helps to activate genes from the virus that allow it to take over the cell. Together, XBP1s and another protein called ATF6 help to switch on the viral genes. The virus also triggers IRE1 helping to reduce the levels of XBP1u, which could slow down the infection. Later, suppressing the unfolded protein response allows copies of the virus to be made faster to help spread the infection. These findings reveal new details of how viruses precisely manipulate their host cells at different stages of infection. These insights could lead to new ways to manage or prevent viral infections.
© 2020, Hinte et al.
Conflict of interest statement
FH, BT, WB No competing interests declared, Ev Reviewing editor, eLife
Figures





Similar articles
-
ZIKV infection activates the IRE1-XBP1 and ATF6 pathways of unfolded protein response in neural cells.J Neuroinflammation. 2018 Sep 21;15(1):275. doi: 10.1186/s12974-018-1311-5. J Neuroinflammation. 2018. PMID: 30241539 Free PMC article.
-
Herpes Simplex Virus 1 UL41 Protein Suppresses the IRE1/XBP1 Signal Pathway of the Unfolded Protein Response via Its RNase Activity.J Virol. 2017 Jan 31;91(4):e02056-16. doi: 10.1128/JVI.02056-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27928013 Free PMC article.
-
[Crosstalk between activating transcription factor 6 and the inositol-requiring enzyme 1-X-box binding protein 1 pathway in oxygen-glucose deprivation/reoxygenation-injured HT22 cells].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2023 Mar;35(3):278-286. doi: 10.3760/cma.j.cn121430-20230228-00115. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2023. PMID: 36916341 Chinese.
-
The molecular mechanism and functional diversity of UPR signaling sensor IRE1.Life Sci. 2021 Jan 15;265:118740. doi: 10.1016/j.lfs.2020.118740. Epub 2020 Nov 11. Life Sci. 2021. PMID: 33188833 Review.
-
Roles of XBP1s in Transcriptional Regulation of Target Genes.Biomedicines. 2021 Jul 8;9(7):791. doi: 10.3390/biomedicines9070791. Biomedicines. 2021. PMID: 34356855 Free PMC article. Review.
Cited by
-
Endoplasmic Reticulum Stress in Cancer.MedComm (2020). 2025 Jun 19;6(7):e70263. doi: 10.1002/mco2.70263. eCollection 2025 Jul. MedComm (2020). 2025. PMID: 40547943 Free PMC article. Review.
-
Viral mediated tethering to SEL1L facilitates ER-associated degradation of IRE1.J Virol. 2021 Mar 25;95(8):e01990-20. doi: 10.1128/JVI.01990-20. Epub 2021 Jan 20. J Virol. 2021. PMID: 33472927 Free PMC article.
-
A cytomegalovirus inflammasome inhibitor reduces proinflammatory cytokine release and pyroptosis.Nat Commun. 2024 Jan 26;15(1):786. doi: 10.1038/s41467-024-45151-z. Nat Commun. 2024. PMID: 38278864 Free PMC article.
-
Herpesviruses and the Unfolded Protein Response.Viruses. 2019 Dec 21;12(1):17. doi: 10.3390/v12010017. Viruses. 2019. PMID: 31877732 Free PMC article. Review.
-
Molecular cloning and host range analysis of three cytomegaloviruses from Mastomys natalensis.J Virol. 2025 May 20;99(5):e0214724. doi: 10.1128/jvi.02147-24. Epub 2025 Apr 9. J Virol. 2025. PMID: 40202317 Free PMC article.
References
-
- Boeck JM, Spencer JV. Cytomegalovirus Encoded G Protein-Coupled Receptors. SM Online Scientific Resources; 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

