Identification of genome-wide single-nucleotide polymorphisms among geographically diverse radish accessions
- PMID: 32065621
- PMCID: PMC7315352
- DOI: 10.1093/dnares/dsaa001
Identification of genome-wide single-nucleotide polymorphisms among geographically diverse radish accessions
Abstract
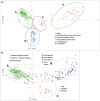
Radish (Raphanus sativus L.) is cultivated around the world as a vegetable crop and exhibits diverse morphological and physiological features. DNA polymorphisms are responsible for differences in traits among cultivars. In this study, we determined genome-wide single-nucleotide polymorphisms (SNPs) among geographically diverse radish accessions using the double-digest restriction site-associated DNA sequencing (ddRAD-Seq) method. A total of 52,559 SNPs was identified in a collection of over 500 radish accessions (cultivated and wild) from East Asia, South and Southeast Asia, and the Occident and Near East. In addition, 2,624 SNP sites without missing data (referred to as common SNP sites) were identified among 510 accessions. Genetic diversity analyses, based on the common SNP sites, divided the cultivated radish accessions into four main groups, each derived from four geographical areas (Japan, East Asia, South and Southeast Asia, and the Occident and Near East). Furthermore, we discuss the origin of cultivated radish and its migration from the West to East Asia. SNP data generated in this work will facilitate further genetic studies on the radish breeding and production of DNA markers.
Keywords: ddRAD-Seq; genome-wide SNPs; radish accessions.
© The Author(s) 2020. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures


Similar articles
-
Comprehensive analysis of expressed sequence tags from cultivated and wild radish (Raphanus spp.).BMC Genomics. 2013 Oct 21;14:721. doi: 10.1186/1471-2164-14-721. BMC Genomics. 2013. PMID: 24144082 Free PMC article.
-
Identification of candidate domestication regions in the radish genome based on high-depth resequencing analysis of 17 genotypes.Theor Appl Genet. 2016 Sep;129(9):1797-814. doi: 10.1007/s00122-016-2741-z. Epub 2016 Jul 4. Theor Appl Genet. 2016. PMID: 27377547
-
Development of SNP and InDel markers by genome resequencing and transcriptome sequencing in radish (Raphanus sativus L.).BMC Genomics. 2023 Aug 8;24(1):445. doi: 10.1186/s12864-023-09528-6. BMC Genomics. 2023. PMID: 37553577 Free PMC article.
-
Identification of highly variable chloroplast sequences and development of cpDNA-based molecular markers that distinguish four cytoplasm types in radish (Raphanus sativus L.).Theor Appl Genet. 2009 Jun;119(1):189-98. doi: 10.1007/s00122-009-1028-z. Epub 2009 Apr 11. Theor Appl Genet. 2009. PMID: 19363601
-
Genetic engineering of radish: current achievements and future goals.Plant Cell Rep. 2011 May;30(5):733-44. doi: 10.1007/s00299-010-0978-6. Epub 2010 Dec 30. Plant Cell Rep. 2011. PMID: 21191596 Review.
Cited by
-
SSR-Sequencing Reveals the Inter- and Intraspecific Genetic Variation and Phylogenetic Relationships among an Extensive Collection of Radish (Raphanus) Germplasm Resources.Biology (Basel). 2021 Nov 30;10(12):1250. doi: 10.3390/biology10121250. Biology (Basel). 2021. PMID: 34943165 Free PMC article.
-
S haplotype collection in Brassicaceae crops-an updated list of S haplotypes.Breed Sci. 2023 Apr;73(2):132-145. doi: 10.1270/jsbbs.22091. Epub 2023 May 17. Breed Sci. 2023. PMID: 37404351 Free PMC article.
-
Genetic Diversity Analysis and Core Germplasm Collection Construction of Radish Cultivars Based on Structure Variation Markers.Int J Mol Sci. 2023 Jan 29;24(3):2554. doi: 10.3390/ijms24032554. Int J Mol Sci. 2023. PMID: 36768875 Free PMC article.
-
Characterization of flavor and taste profile of different radish (Raphanus Sativus L.) varieties by headspace-gas chromatography-ion mobility spectrometry (GC/IMS) and E-nose/tongue.Food Chem X. 2024 Apr 30;22:101419. doi: 10.1016/j.fochx.2024.101419. eCollection 2024 Jun 30. Food Chem X. 2024. PMID: 38756475 Free PMC article.
-
Sequence and epigenetic variations of R2R3-MYB transcription factors determine the diversity of taproot skin and flesh colors in different cultivated types of radish (Raphanus sativus L.).Theor Appl Genet. 2024 May 16;137(6):133. doi: 10.1007/s00122-024-04631-y. Theor Appl Genet. 2024. PMID: 38753199
References
-
- Jeong Y.M., Kim N., Ahn B.O., et al.2016, Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes, Theor. Appl. Genet., 129, 1357–72. - PubMed
-
- Zhang X., Yue Z., Mei S., et al.2015, A de novo genome of a Chinese Radish cultivar, Horticul. Plant J., 1, 155–64.
-
- Shirasawa K., Kitashiba H.. 2017, Genetic maps and whole genome sequences of Radish, In: Nishio T., Kitashiba H. (eds) The Radish Genome, Springer International Publishing, Switzerland, pp. 31–42.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

