The complete mitochondrial genome of a parasite at the animal-fungal boundary
- PMID: 32066491
- PMCID: PMC7027106
- DOI: 10.1186/s13071-020-3926-5
The complete mitochondrial genome of a parasite at the animal-fungal boundary
Abstract
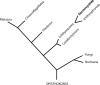
Background: Sphaerothecum destruens is an obligate intracellular fish parasite which has been identified as a serious threat to freshwater fishes. Taxonomically, S. destruens belongs to the order Dermocystida within the class Ichthyosporea (formerly referred to as Mesomycetozoea), which sits at the animal-fungal boundary. Mitochondrial DNA (mtDNA) sequences can be valuable genetic markers for species detection and are increasingly used in environmental DNA (eDNA) based species detection. Furthermore, mtDNA sequences can be used in epidemiological studies by informing detection, strain identification and geographical spread.
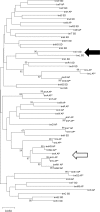
Methods: We amplified the entire mitochondrial (mt) genome of S. destruens in two overlapping long fragments using primers designed based on the cox1, cob and nad5 partial sequences. The mt-genome architecture of S. destruens was then compared to close relatives to gain insights into its evolution.
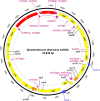
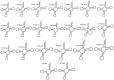
Results: The complete mt-genome of Sphaerothecum destruens is 23,939 bp in length and consists of 47 genes including 21 protein-coding genes, 2 rRNA, 22 tRNA and two unidentified open reading frames. The mitochondrial genome of S. destruens is intronless and compact with a few intergenic regions and includes genes that are often missing from animal and fungal mt-genomes, such as, the four ribosomal proteins (small subunit rps13 and 14; large subunit rpl2 and 16), tatC (twin-arginine translocase component C), and ccmC and ccmF (cytochrome c maturation protein ccmC and heme lyase).
Conclusions: We present the first mt-genome of S. destruens which also represents the first mt-genome for the order Dermocystida. The availability of the mt-genome can assist the detection of S. destruens and closely related parasites in eukaryotic diversity surveys using eDNA and assist epidemiological studies by improving molecular detection and tracking the parasite's spread. Furthermore, as the only representative of the order Dermocystida, its mt-genome can be used in the study of mitochondrial evolution of the unicellular relatives of animals.
Keywords: Animal-fungal boundary; Dermocystida; Invasive; Mesomycetozoea; Mitochondrial DNA; Parasite; Pseudorasbora parva; Sphaerothecum destruens; Topmouth gudgeon.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

