HIV-1 gp120-CD4-Induced Antibody Complex Elicits CD4 Binding Site-Specific Antibody Response in Mice
- PMID: 32066595
- PMCID: PMC7065964
- DOI: 10.4049/jimmunol.1901051
HIV-1 gp120-CD4-Induced Antibody Complex Elicits CD4 Binding Site-Specific Antibody Response in Mice
Abstract
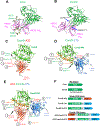
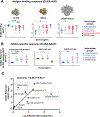
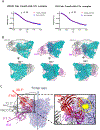
Elicitation of broadly neutralizing Ab (bNAb) responses toward the conserved HIV-1 envelope (Env) CD4 binding site (CD4bs) by vaccination is an important goal for vaccine development and yet to be achieved. The outcome of previous immunogenicity studies suggests that the limited accessibility of the CD4bs and the presence of predominant nonneutralizing determinants (nND) on Env may impede the elicitation of bNAbs and their precursors by vaccination. In this study, we designed a panel of novel immunogens that 1) preferentially expose the CD4bs by selective elimination of glycosylation sites flanking the CD4bs, and 2) minimize the nND immune response by engineering fusion proteins consisting of gp120 Core and one or two CD4-induced (CD4i) mAbs for masking nND epitopes, referred to as gp120-CD4i fusion proteins. As expected, the fusion proteins possess improved antigenicity with retained affinity for VRC01-class, CD4bs-directed bNAbs and dampened affinity for nonneutralizing Abs. We immunized C57BL/6 mice with these fusion proteins and found that overall the fusion proteins elicit more focused CD4bs Ab response than prototypical gp120 Core by serological analysis. Consistently, we found that mice immunized with selected gp120-CD4i fusion proteins have higher frequencies of germinal center-activated B cells and CD4bs-directed memory B cells than those inoculated with parental immunogens. We isolated three mAbs from mice immunized with selected gp120-CD4i fusion proteins and found that their footprints on Env are similar to VRC01-class bNAbs. Thus, using gp120-CD4i fusion proteins with selective glycan deletion as immunogens could focus Ab response toward CD4bs epitope.
Copyright © 2020 by The American Association of Immunologists, Inc.
Conflict of interest statement
Figures








References
-
- Wyatt R, and Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280: 1884–1888. - PubMed
-
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, and Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384: 179–183. - PubMed
-
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, and Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514: 455–461. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

