FOXM1 regulates leukemia stem cell quiescence and survival in MLL-rearranged AML
- PMID: 32066721
- PMCID: PMC7026046
- DOI: 10.1038/s41467-020-14590-9
FOXM1 regulates leukemia stem cell quiescence and survival in MLL-rearranged AML
Abstract
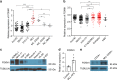
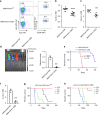
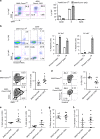
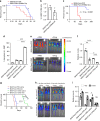
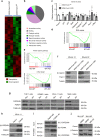
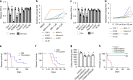
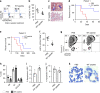
FOXM1, a known transcription factor, promotes cell proliferation in a variety of cancer cells. Here we show that Foxm1 is required for survival, quiescence and self-renewal of MLL-AF9 (MA9)-transformed leukemia stem cells (LSCs) in vivo. Mechanistically, Foxm1 upregulation activates the Wnt/β-catenin signaling pathways by directly binding to β-catenin and stabilizing β-catenin protein through inhibiting its degradation, thereby preserving LSC quiescence, and promoting LSC self-renewal in MLL-rearranged AML. More importantly, inhibition of FOXM1 markedly suppresses leukemogenic potential and induces apoptosis of primary LSCs from MLL-rearranged AML patients in vitro and in vivo in xenograft mice. Thus, our study shows a critical role and mechanisms of Foxm1 in MA9-LSCs, and indicates that FOXM1 is a potential therapeutic target for selectively eliminating LSCs in MLL-rearranged AML.
Conflict of interest statement
The authors declare no competing interests
Figures


Similar articles
-
PBX3 is essential for leukemia stem cell maintenance in MLL-rearranged leukemia.Int J Cancer. 2017 Jul 15;141(2):324-335. doi: 10.1002/ijc.30739. Epub 2017 May 8. Int J Cancer. 2017. PMID: 28411381
-
Blast cells surviving acute myeloid leukemia induction therapy are in cycle with a signature of FOXM1 activity.BMC Cancer. 2021 Oct 28;21(1):1153. doi: 10.1186/s12885-021-08839-9. BMC Cancer. 2021. PMID: 34711181 Free PMC article.
-
Bortezomib suppresses self-renewal and leukemogenesis of leukemia stem cell by NF-ĸB-dependent inhibition of CDK6 in MLL-rearranged myeloid leukemia.J Cell Mol Med. 2021 Mar;25(6):3124-3135. doi: 10.1111/jcmm.16377. Epub 2021 Feb 17. J Cell Mol Med. 2021. PMID: 33599085 Free PMC article.
-
Selective treatment of mixed-lineage leukemia leukemic stem cells through targeting glycogen synthase kinase 3 and the canonical Wnt/β-catenin pathway.Curr Opin Hematol. 2012 Jul;19(4):280-6. doi: 10.1097/MOH.0b013e3283545615. Curr Opin Hematol. 2012. PMID: 22525581 Review.
-
Regulation of the master regulator FOXM1 in cancer.Cell Commun Signal. 2018 Sep 12;16(1):57. doi: 10.1186/s12964-018-0266-6. Cell Commun Signal. 2018. PMID: 30208972 Free PMC article. Review.
Cited by
-
USP39 facilitates breast cancer cell proliferation through stabilization of FOXM1.Am J Cancer Res. 2022 Aug 15;12(8):3644-3661. eCollection 2022. Am J Cancer Res. 2022. PMID: 36119839 Free PMC article.
-
Impairment of FOXM1 expression in mesenchymal cells from patients with myeloid neoplasms, de novo and therapy-related, may compromise their ability to support hematopoiesis.Sci Rep. 2022 Dec 8;12(1):21231. doi: 10.1038/s41598-022-24644-1. Sci Rep. 2022. PMID: 36481766 Free PMC article.
-
Cell-intrinsic factors governing quiescence vis-à-vis activation of adult hematopoietic stem cells.Mol Cell Biochem. 2023 Jun;478(6):1361-1382. doi: 10.1007/s11010-022-04594-y. Epub 2022 Oct 30. Mol Cell Biochem. 2023. PMID: 36309884 Review.
-
Distinctive molecular features of regenerative stem cells in the damaged male germline.Nat Commun. 2022 May 6;13(1):2500. doi: 10.1038/s41467-022-30130-z. Nat Commun. 2022. PMID: 35523793 Free PMC article.
-
Alkynyl nicotinamides show antileukemic activity in drug-resistant acute myeloid leukemia.J Clin Invest. 2024 Jun 17;134(12):e169245. doi: 10.1172/JCI169245. J Clin Invest. 2024. PMID: 38950330 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

