Distinct microbial and immune niches of the human colon
- PMID: 32066951
- PMCID: PMC7212050
- DOI: 10.1038/s41590-020-0602-z
Distinct microbial and immune niches of the human colon
Abstract
Gastrointestinal microbiota and immune cells interact closely and display regional specificity; however, little is known about how these communities differ with location. Here, we simultaneously assess microbiota and single immune cells across the healthy, adult human colon, with paired characterization of immune cells in the mesenteric lymph nodes, to delineate colonic immune niches at steady state. We describe distinct helper T cell activation and migration profiles along the colon and characterize the transcriptional adaptation trajectory of regulatory T cells between lymphoid tissue and colon. Finally, we show increasing B cell accumulation, clonal expansion and mutational frequency from the cecum to the sigmoid colon and link this to the increasing number of reactive bacterial species.
Conflict of interest statement
S.C.F. and T.D.L. are either employees of, or consultants to, Microbiotica Pty Ltd.
Figures
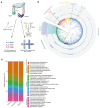

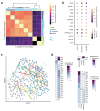


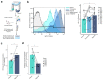
Comment in
-
Colons or semi-colons: punctuating the regional variation of intestinal microbial-immune interactions.Nat Rev Gastroenterol Hepatol. 2020 Jun;17(6):319-320. doi: 10.1038/s41575-020-0302-z. Nat Rev Gastroenterol Hepatol. 2020. PMID: 32322050 No abstract available.
References
-
- Wang X, Heazlewood SP, Krause DO, Florin THJ. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol. 2003;95:508–520. - PubMed
-
- Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

