CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors
- PMID: 32066953
- PMCID: PMC7043937
- DOI: 10.1038/s41590-019-0589-5
CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors
Abstract
Depleting regulatory T cells (Treg cells) to counteract immunosuppressive features of the tumor microenvironment (TME) is an attractive strategy for cancer treatment; however, autoimmunity due to systemic impairment of their suppressive function limits its therapeutic potential. Elucidating approaches that specifically disrupt intratumoral Treg cells is direly needed for cancer immunotherapy. We found that CD36 was selectively upregulated in intrautumoral Treg cells as a central metabolic modulator. CD36 fine-tuned mitochondrial fitness via peroxisome proliferator-activated receptor-β signaling, programming Treg cells to adapt to a lactic acid-enriched TME. Genetic ablation of Cd36 in Treg cells suppressed tumor growth accompanied by a decrease in intratumoral Treg cells and enhancement of antitumor activity in tumor-infiltrating lymphocytes without disrupting immune homeostasis. Furthermore, CD36 targeting elicited additive antitumor responses with anti-programmed cell death protein 1 therapy. Our findings uncover the unexplored metabolic adaptation that orchestrates the survival and functions of intratumoral Treg cells, and the therapeutic potential of targeting this pathway for reprogramming the TME.
Conflict of interest statement
H.W. and P.-C.H. are inventors of patent application related to targeting CD36 in cancer immunotherapy. P.-C.H. is serving as a member of scientific advisory board for Elixiron Immunotherapeutics and receiving research grants from Roche and Idorsia. J.D.W. is serving as a consultant for Adaptive Biotech; Advaxis; Amgen; Apricity; Array BioPharma; Ascentage Pharma; Astellas; Bayer; Beigene; Bristol Myers Squibb; Celgene; Chugai; Elucida; Eli Lilly; F Star; Genentech; Imvaq; Janssen; Kyowa Hakko Kirin; Kleo Pharma; Linneaus;MedImmune; Merck; Neon Therapuetics; Northern Biologics; Ono; Polaris Pharma; Polynoma; Psioxus; Puretech; Recepta; Takara Bio; Trieza; Sellas Life Sciences; Serametrix; Surface Oncology; Syndax; Synthologic. J.D.W. received research support from Bristol Myers Squibb; Medimmune; Merck Pharmaceuticals; Genentech and has equity in Potenza Therapeutics; Tizona Pharmaceuticals; Adaptive Biotechnologies; Elucida; Imvaq; Beigene; Trieza; Linneaus. P.C.H. received honorarium from Pfizer and Chugai and J.D.W received honorarium from Esanex.
Figures

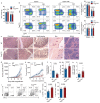

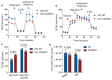
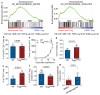


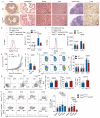



Comment in
-
CD36 - the Achilles' heel of Treg cells.Nat Immunol. 2020 Mar;21(3):251-253. doi: 10.1038/s41590-020-0601-0. Nat Immunol. 2020. PMID: 32066948 No abstract available.
References
-
- Sutmuller RP, et al. Synergism of cytotoxic T lymphocyte-associated antigen 4 blockade and depletion of CD25(+) regulatory T cells in antitumor therapy reveals alternative pathways for suppression of autoreactive cytotoxic T lymphocyte responses. J Exp Med. 2001;194:823–832. doi: 10.1084/jem.194.6.823. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

