FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair
- PMID: 32066963
- PMCID: PMC7067600
- DOI: 10.1038/s41594-020-0380-1
FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair
Abstract
Vertebrate DNA crosslink repair excises toxic replication-blocking DNA crosslinks. Numerous factors involved in crosslink repair have been identified, and mutations in their corresponding genes cause Fanconi anemia (FA). A key step in crosslink repair is monoubiquitination of the FANCD2-FANCI heterodimer, which then recruits nucleases to remove the DNA lesion. Here, we use cryo-EM to determine the structures of recombinant chicken FANCD2 and FANCI complexes. FANCD2-FANCI adopts a closed conformation when the FANCD2 subunit is monoubiquitinated, creating a channel that encloses double-stranded DNA (dsDNA). Ubiquitin is positioned at the interface of FANCD2 and FANCI, where it acts as a covalent molecular pin to trap the complex on DNA. In contrast, isolated FANCD2 is a homodimer that is unable to bind DNA, suggestive of an autoinhibitory mechanism that prevents premature activation. Together, our work suggests that FANCD2-FANCI is a clamp that is locked onto DNA by ubiquitin, with distinct interfaces that may recruit other DNA repair factors.
Conflict of interest statement
Figures


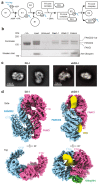



Comment in
-
Ubiquitilated Fanconi ID complex embraces DNA.Cell Res. 2020 Jul;30(7):554-555. doi: 10.1038/s41422-020-0345-2. Cell Res. 2020. PMID: 32472046 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

