A native function for RAN translation and CGG repeats in regulating fragile X protein synthesis
- PMID: 32066985
- PMCID: PMC7668390
- DOI: 10.1038/s41593-020-0590-1
A native function for RAN translation and CGG repeats in regulating fragile X protein synthesis
Abstract
Repeat-associated non-AUG-initiated translation of expanded CGG repeats (CGG RAN) from the FMR1 5'-leader produces toxic proteins that contribute to neurodegeneration in fragile X-associated tremor/ataxia syndrome. Here we describe how unexpanded CGG repeats and their translation play conserved roles in regulating fragile X protein (FMRP) synthesis. In neurons, CGG RAN acts as an inhibitory upstream open reading frame to suppress basal FMRP production. Activation of mGluR5 receptors enhances FMRP synthesis. This enhancement requires both the CGG repeat and CGG RAN initiation sites. Using non-cleaving antisense oligonucleotides (ASOs), we selectively blocked CGG RAN. This ASO blockade enhanced endogenous FMRP expression in human neurons. In human and rodent neurons, CGG RAN-blocking ASOs suppressed repeat toxicity and prolonged survival. These findings delineate a native function for CGG repeats and RAN translation in regulating basal and activity-dependent FMRP synthesis, and they demonstrate the therapeutic potential of modulating CGG RAN translation in fragile X-associated disorders.
Conflict of interest statement
Competing interests
P.K.T. served as a consultant with Denali Therapeutics and has licensed technology through the University of Michigan to Denali. P.K.T., C.M.R, F.R., and P.J. hold a patent on ASOs targeting CGG RAN. F.R. and P.J. are paid employees of Ionis Pharmaceuticals. All other authors declare no competing interests.
Figures
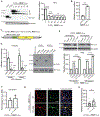







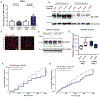

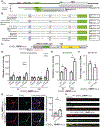
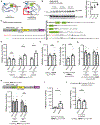

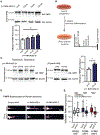
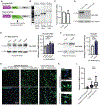
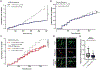
References
-
- Paulson H in Handbook of clinical neurology Vol. 147 (eds Geschwind Daniel H., Paulson Henry L., & Klein Christine) 105–123 (Elsevier, 2018).
-
- Verkerk AJ et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914 (1991). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

