Single-cell analysis of human adipose tissue identifies depot and disease specific cell types
- PMID: 32066997
- PMCID: PMC7025882
- DOI: 10.1038/s42255-019-0152-6
Single-cell analysis of human adipose tissue identifies depot and disease specific cell types
Abstract
The complex relationship between metabolic disease risk and body fat distribution in humans involves cellular characteristics which are specific to body fat compartments. Here we show depot-specific differences in the stromal vascual fraction of visceral and subcutaneous adipose tissue by performing single-cell RNA sequencing of tissue specimen from obese individuals. We characterize multiple immune cells, endothelial cells, fibroblasts, adipose and hematopoietic stem cell progenitors. Subpopulations of adipose-resident immune cells are metabolically active and associated with metabolic disease status and those include a population of potential dysfunctional CD8+ T cells expressing metallothioneins. We identify multiple types of adipocyte progenitors that are common across depots, including a subtype enriched in individuals with type 2 diabetes. Depot-specific analysis reveals a class of adipocyte progenitors unique to visceral adipose tissue, which shares common features with beige preadipocytes. Our human single-cell transcriptome atlas across fat depots provides a resource to dissect functional genomics of metabolic disease.
Conflict of interest statement
Declaration of Interests A.T. receives funding from Johnson & Johnson Medical Companies and Medtronic for research unrelated to the present manuscript.
Figures


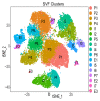
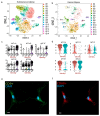

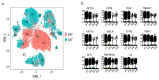
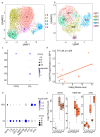


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

