A widely distributed metalloenzyme class enables gut microbial metabolism of host- and diet-derived catechols
- PMID: 32067637
- PMCID: PMC7028382
- DOI: 10.7554/eLife.50845
A widely distributed metalloenzyme class enables gut microbial metabolism of host- and diet-derived catechols
Abstract
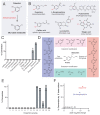
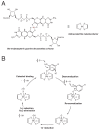
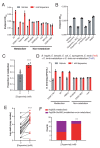
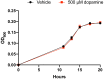
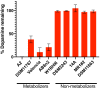
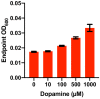
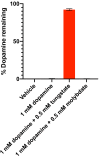
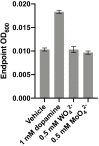
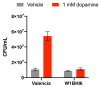
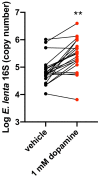
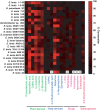
Catechol dehydroxylation is a central chemical transformation in the gut microbial metabolism of plant- and host-derived small molecules. However, the molecular basis for this transformation and its distribution among gut microorganisms are poorly understood. Here, we characterize a molybdenum-dependent enzyme from the human gut bacterium Eggerthella lenta that dehydroxylates catecholamine neurotransmitters. Our findings suggest that this activity enables E. lenta to use dopamine as an electron acceptor. We also identify candidate dehydroxylases that metabolize additional host- and plant-derived catechols. These dehydroxylases belong to a distinct group of largely uncharacterized molybdenum-dependent enzymes that likely mediate primary and secondary metabolism in multiple environments. Finally, we observe catechol dehydroxylation in the gut microbiotas of diverse mammals, confirming the presence of this chemistry in habitats beyond the human gut. These results suggest that the chemical strategies that mediate metabolism and interactions in the human gut are relevant to a broad range of species and habitats.
Keywords: Eggerthella lenta; Gordonibacter sp.; biochemistry; catechol dehydroxylase; chemical biology; infectious disease; microbiology; molybdenum enzyme.
Plain language summary
Inside the human gut there are trillions of bacteria. These microbes are critical for breaking down and modifying molecules that the body consumes (such as nutrients and drugs) and produces (such as hormones). Although metabolizing these molecules is known to impact health and disease, little is known about the specific components, such as the genes and enzymes, involved in these reactions. A prominent microbial reaction in the gut metabolizes molecules by removing a hydroxyl group from an aromatic ring and replacing it with a hydrogen atom. This chemical reaction influences the fate of dietary compounds, clinically used drugs and chemicals which transmit signals between nerves (neurotransmitters). But even though this reaction was discovered over 50 years ago, it remained unknown which microbial enzymes are directly responsible for this metabolism. In 2019, researchers discovered the human gut bacteria Eggerthella lenta produces an enzyme named Dadh that can remove a hydroxyl group from the neurotransmitter dopamine. Now, Maini Rekdal et al. – including many of the researchers involved in the 2019 study – have used a range of different experiments to further characterize this enzyme and see if it can break down molecules other than dopamine. This revealed that Dadh specifically degrades dopamine, and this process promotes E. lenta growth. Next, Maini Rekdal et al. uncovered a group of enzymes that had similar characteristics to Dadh and could metabolize molecules other than dopamine, including molecules derived from plants and nutrients in food. These Dadh-like enzymes were found not only in the guts of humans, but in other organisms and environments, including the soil, ocean and plants. Plant-derived molecules are associated with human health, and the discovery of the enzymes that break down these products could provide new insights into the health effects of plant-based foods. In addition, the finding that gut bacteria harbor a dopamine metabolizing enzyme has implications for the interaction between the gut microbiome and the nervous system, which has been linked to human health and disease. These newly discovered enzymes are also involved in metabolic reactions outside the human body. Future work investigating the mechanisms and outputs of these reactions could improve current strategies for degrading pollutants and producing medically useful molecules.
© 2020, Maini Rekdal et al.
Conflict of interest statement
VM, PN, ML, SK, CL, JB, EB No competing interests declared, PT Reviewing editor, eLife, EB has consulted for Merck, Novartis, and Kintai Therapeutics; is on the Scientific Advisory Boards of Kintai Therapeutics and Caribou Biosciences.
Figures









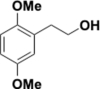
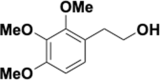
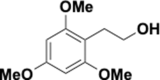

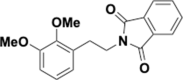
Similar articles
-
Metatranscriptomics-guided discovery and characterization of a polyphenol-metabolizing gut microbial enzyme.Cell Host Microbe. 2024 Nov 13;32(11):1887-1896.e8. doi: 10.1016/j.chom.2024.10.002. Epub 2024 Oct 28. Cell Host Microbe. 2024. PMID: 39471822 Free PMC article.
-
Enantiocomplementary Gut Bacterial Enzymes Metabolize Dietary Polyphenols.J Am Chem Soc. 2025 Mar 5;147(9):7231-7244. doi: 10.1021/jacs.4c09892. Epub 2025 Feb 24. J Am Chem Soc. 2025. PMID: 39993729 Free PMC article.
-
Host-microbiome metabolism of a plant toxin in bees.Elife. 2022 Dec 6;11:e82595. doi: 10.7554/eLife.82595. Elife. 2022. PMID: 36472498 Free PMC article.
-
Iron-sulfur cluster-dependent enzymes and molybdenum-dependent reductases in the anaerobic metabolism of human gut microbes.Metallomics. 2024 Nov 7;16(11):mfae049. doi: 10.1093/mtomcs/mfae049. Metallomics. 2024. PMID: 39504489 Free PMC article. Review.
-
Diverse mechanisms by which chemical pollutant exposure alters gut microbiota metabolism and inflammation.Environ Int. 2024 Aug;190:108805. doi: 10.1016/j.envint.2024.108805. Epub 2024 Jun 10. Environ Int. 2024. PMID: 38901183 Free PMC article. Review.
Cited by
-
Challenges and opportunities of strain diversity in gut microbiome research.Front Microbiol. 2023 Feb 17;14:1117122. doi: 10.3389/fmicb.2023.1117122. eCollection 2023. Front Microbiol. 2023. PMID: 36876113 Free PMC article. Review.
-
The microbiota-gut-brain axis: pathways to better brain health. Perspectives on what we know, what we need to investigate and how to put knowledge into practice.Cell Mol Life Sci. 2022 Jan 19;79(2):80. doi: 10.1007/s00018-021-04060-w. Cell Mol Life Sci. 2022. PMID: 35044528 Free PMC article. Review.
-
Conceptual Exchanges for Understanding Free-Living and Host-Associated Microbiomes.mSystems. 2022 Feb 22;7(1):e0137421. doi: 10.1128/msystems.01374-21. Epub 2022 Jan 11. mSystems. 2022. PMID: 35014872 Free PMC article.
-
Polyphenol rewiring of the microbiome reduces methane emissions.ISME J. 2025 Jan 2;19(1):wraf108. doi: 10.1093/ismejo/wraf108. ISME J. 2025. PMID: 40439232 Free PMC article.
-
Metatranscriptomics-guided discovery and characterization of a polyphenol-metabolizing gut microbial enzyme.Cell Host Microbe. 2024 Nov 13;32(11):1887-1896.e8. doi: 10.1016/j.chom.2024.10.002. Epub 2024 Oct 28. Cell Host Microbe. 2024. PMID: 39471822 Free PMC article.
References
-
- Arnow LE. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine tyrosine mixtures. The Journal of Biological Chemistry. 1937;118:531–537.
-
- Aura A-M, Mattila I, Seppänen-Laakso T, Miettinen J, Oksman-Caldentey K-M, Orešič M. Microbial metabolism of catechin stereoisomers by human faecal microbiota: Comparison of targeted analysis and a non-targeted metabolomics method. Phytochemistry Letters. 2008;1:18–22. doi: 10.1016/j.phytol.2007.12.001. - DOI
-
- Bess EN, Bisanz JE, Yarza F, Bustion A, Rich BE, Li X, Kitamura S, Waligurski E, Ang QY, Alba DL, Spanogiannopoulos P, Nayfach S, Koliwad SK, Wolan DW, Franke AA, Turnbaugh PJ. Genetic basis for the cooperative bioactivation of plant lignans by Eggerthella lenta and other human gut Bacteria. Nature Microbiology. 2020;5:56–66. doi: 10.1038/s41564-019-0596-1. - DOI - PMC - PubMed
-
- Bisanz JE, Soto-Perez P, Lam KN, Bess EN, Haiser HJ, Allen-Vercoe E, Rekdal VM, Balskus EP, Turnbaugh PJ. Illuminating the microbiome's dark matter: a functional genomic toolkit for the study of human gut Actinobacteria. bioRxiv. 2018 doi: 10.1101/304840. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

