Inferring the Demographic History of Inbred Species from Genome-Wide SNP Frequency Data
- PMID: 32068861
- PMCID: PMC7828618
- DOI: 10.1093/molbev/msaa042
Inferring the Demographic History of Inbred Species from Genome-Wide SNP Frequency Data
Abstract
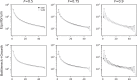
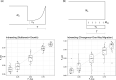
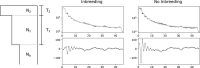
Demographic inference using the site frequency spectrum (SFS) is a common way to understand historical events affecting genetic variation. However, most methods for estimating demography from the SFS assume random mating within populations, precluding these types of analyses in inbred populations. To address this issue, we developed a model for the expected SFS that includes inbreeding by parameterizing individual genotypes using beta-binomial distributions. We then take the convolution of these genotype probabilities to calculate the expected frequency of biallelic variants in the population. Using simulations, we evaluated the model's ability to coestimate demography and inbreeding using one- and two-population models across a range of inbreeding levels. We also applied our method to two empirical examples, American pumas (Puma concolor) and domesticated cabbage (Brassica oleracea var. capitata), inferring models both with and without inbreeding to compare parameter estimates and model fit. Our simulations showed that we are able to accurately coestimate demographic parameters and inbreeding even for highly inbred populations (F = 0.9). In contrast, failing to include inbreeding generally resulted in inaccurate parameter estimates in simulated data and led to poor model fit in our empirical analyses. These results show that inbreeding can have a strong effect on demographic inference, a pattern that was especially noticeable for parameters involving changes in population size. Given the importance of these estimates for informing practices in conservation, agriculture, and elsewhere, our method provides an important advancement for accurately estimating the demographic histories of these species.
Keywords: conservation; demography; domestication; inbreeding; site frequency spectrum.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Balding DJ, Nichols RA.. 1995. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96(1–2):3–12. - PubMed
-
- Balding DJ, Nichols RA.. 1997. Significant genetic correlations among Caucasians at forensic DNA loci. Heredity 108:583–589. - PubMed
-
- Beissinger TM, Wang L, Crosby K, Durvasula A, Hufford MB, Ross-Ibarra J.. 2016. Recent demography drives changes in linked selection across the maize genome. Nat Plants. 2(7):16084. - PubMed
-
- Belser C, Istace B, Denis E, Dubarry M, Baurens F-C, Falentin C, Genete M, Berrabah W, Chèvre A-M, Delourme R, et al. 2018. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat Plants. 4(11):879–887. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

