Genome-Wide Analysis of Gene Expression Provides New Insights into Waterlogging Responses in Barley (Hordeum vulgare L.)
- PMID: 32069892
- PMCID: PMC7076447
- DOI: 10.3390/plants9020240
Genome-Wide Analysis of Gene Expression Provides New Insights into Waterlogging Responses in Barley (Hordeum vulgare L.)
Abstract
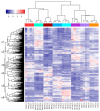
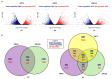
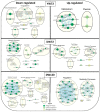
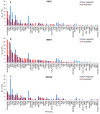
Waterlogging is a major abiotic stress causing oxygen depletion and carbon dioxide accumulation in the rhizosphere. Barley is more susceptible to waterlogging stress than other cereals. To gain a better understanding, the genome-wide gene expression responses in roots of waterlogged barley seedlings of Yerong and Deder2 were analyzed by RNA-Sequencing. A total of 6736, 5482, and 4538 differentially expressed genes (DEGs) were identified in waterlogged roots of Yerong at 72 h and Deder2 at 72 and 120 h, respectively, compared with the non-waterlogged control. Gene Ontology (GO) enrichment analyses showed that the most significant changes in GO terms, resulted from these DEGs observed under waterlogging stress, were related to primary and secondary metabolism, regulation, and oxygen carrier activity. In addition, more than 297 transcription factors, including members of MYB, AP2/EREBP, NAC, WRKY, bHLH, bZIP, and G2-like families, were identified as waterlogging responsive. Tentative important contributors to waterlogging tolerance in Deder2 might be the highest up-regulated DEGs: Trichome birefringence, α/β-Hydrolases, Xylanase inhibitor, MATE efflux, serine carboxypeptidase, and SAUR-like auxin-responsive protein. The study provides insights into the molecular mechanisms underlying the response to waterlogging in barley, which will be of benefit for future studies of molecular responses to waterlogging and will greatly assist barley genetic research and breeding.
Keywords: RNA-Seq; barley; differentially expressed genes; transcription factors; waterlogging stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- FAOSTAT Production. [(accessed on 18 November 2019)]; Available online: http://www.fao.org/faostat/en/#data/QC.
-
- Statistics Canada Estimated areas, yield, production, average farm price and total farm value of principal field crops. [(accessed on 18 November 2019)]; Available online: https://www150.statcan.gc.ca/t1/tbl1/en/tv.action?pid=3210035901.
-
- Canadian Agri-Food Trade Alliance (CAFTA) [(accessed on 18 November 2019)]; Available online: http://cafta.org/agri-food-exports/cafta-exports/
-
- Setter T.L., Waters I. Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil. 2003;253:1–34. doi: 10.1023/A:1024573305997. - DOI
-
- Pang J., Zhou M., Mendham N., Shabala S. Growth and physiological responses of six barley genotypes to waterlogging and subsequent recovery. Aust. J. Agric. Res. 2004;55:895–906. doi: 10.1071/AR03097. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

