Twenty Years of Progress in Physiological and Biochemical Investigation of RALF Peptides
- PMID: 32071151
- PMCID: PMC7140910
- DOI: 10.1104/pp.19.01310
Twenty Years of Progress in Physiological and Biochemical Investigation of RALF Peptides
Abstract
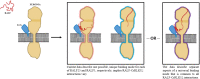
RALF isoforms play many biological roles, and their specific functions are defined by combinatorial interactions with dynamic receptor complexes that vary more than initially thought.
Figures



References
-
- Belkhadir Y, Jaillais Y(2015) The molecular circuitry of brassinosteroid signaling. New Phytol 206: 522–540 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

