CoMPARA: Collaborative Modeling Project for Androgen Receptor Activity
- PMID: 32074470
- PMCID: PMC7064318
- DOI: 10.1289/EHP5580
CoMPARA: Collaborative Modeling Project for Androgen Receptor Activity
Abstract
Background: Endocrine disrupting chemicals (EDCs) are xenobiotics that mimic the interaction of natural hormones and alter synthesis, transport, or metabolic pathways. The prospect of EDCs causing adverse health effects in humans and wildlife has led to the development of scientific and regulatory approaches for evaluating bioactivity. This need is being addressed using high-throughput screening (HTS) in vitro approaches and computational modeling.
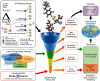
Objectives: In support of the Endocrine Disruptor Screening Program, the U.S. Environmental Protection Agency (EPA) led two worldwide consortiums to virtually screen chemicals for their potential estrogenic and androgenic activities. Here, we describe the Collaborative Modeling Project for Androgen Receptor Activity (CoMPARA) efforts, which follows the steps of the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP).
Methods: The CoMPARA list of screened chemicals built on CERAPP's list of 32,464 chemicals to include additional chemicals of interest, as well as simulated ToxCast™ metabolites, totaling 55,450 chemical structures. Computational toxicology scientists from 25 international groups contributed 91 predictive models for binding, agonist, and antagonist activity predictions. Models were underpinned by a common training set of 1,746 chemicals compiled from a combined data set of 11 ToxCast™/Tox21 HTS in vitro assays.
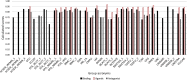
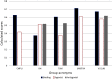
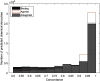
Results: The resulting models were evaluated using curated literature data extracted from different sources. To overcome the limitations of single-model approaches, CoMPARA predictions were combined into consensus models that provided averaged predictive accuracy of approximately 80% for the evaluation set.
Discussion: The strengths and limitations of the consensus predictions were discussed with example chemicals; then, the models were implemented into the free and open-source OPERA application to enable screening of new chemicals with a defined applicability domain and accuracy assessment. This implementation was used to screen the entire EPA DSSTox database of chemicals, and their predicted AR activities have been made available on the EPA CompTox Chemicals dashboard and National Toxicology Program's Integrated Chemical Environment. https://doi.org/10.1289/EHP5580.
Figures

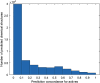
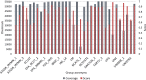
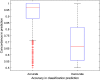
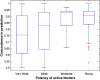

References
-
- Ballabio D, Grisoni F, Todeschini R. 2018. Multivariate comparison of classification performance measures. Chemometr Intell Lab Syst 174:33–44, 10.1016/j.chemolab.2017.12.004. - DOI
-
- Ballabio D, Vasighi M, Consonni V, Kompany-Zareh M. 2011. Genetic algorithms for architecture optimisation of counter-propagation artificial neural networks. Chemometr Intell Lab Syst 105(1):56–64, 10.1016/j.chemolab.2010.10.010. - DOI
-
- Benigni R. 2003. Quantitative Structure-Activity Relationship (QSAR) Models of Mutagens and Carcinogens. Boca Raton, FL: CRC Press. - PubMed
-
- Berk RA. 2008. Statistical Learning from a Regression Perspective. New York, NY: Springer-Verlag.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
