ZnO Nanolower-Based NanoPCR as an Efficient Diagnostic Tool for Quick Diagnosis of Canine Vector-Borne Pathogens
- PMID: 32075178
- PMCID: PMC7169380
- DOI: 10.3390/pathogens9020122
ZnO Nanolower-Based NanoPCR as an Efficient Diagnostic Tool for Quick Diagnosis of Canine Vector-Borne Pathogens
Abstract
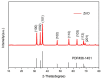
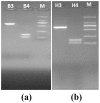
: Polymerase chain reaction (PCR) is a unique technique in molecular biology and biotechnology for amplifying target DNA strands, and is also considered as a gold standard for the diagnosis of many canine diseases as well as many other infectious diseases. However, PCR still faces many challenges and issues related to its sensitivity, specificity, efficiency, and turnaround time. To address these issues, we described the use of unique ZnO nanoflowers in PCR reaction and an efficient ZnO nanoflower-based PCR (nanoPCR) for the molecular diagnosis of canine vector-borne diseases (CVBDs). A total of 1 mM of an aqueous solution of ZnO nanoflowers incorporated in PCR showed a significant enhancement of the PCR assay with respect to its sensitivity and specificity for the diagnosis of two important CVBDs, Babesia canis vogeli and Hepatozoon canis. Interestingly, it drastically reduced the turnaround time of the PCR assay without compromising the yield of the amplified DNA, which can be of benefit for veterinary practitioners for the improved management of diseases. This can be attributed to the favorable adsorption of ZnO nanoflowers to the DNA and thermal conductivity of ZnO nanoflowers. The unique ZnO nanoflower-assisted nanoPCR greatly improved the yield, purity, and quality of the amplified products, but the mechanism behind these properties and the effects and changes due to the different concentrations of ZnO nanoflowers in the PCR system needs to be further studied.
Keywords: Babesia canis vogeli; Hepatozoon canis; PCR; ZnO nanoflowers; canine vector-borne diseases; nanoPCR; nanomaterial-assisted polymerase chain reaction.
Conflict of interest statement
There were no competing interests.
Figures


Similar articles
-
A new PCR assay for the detection and differentiation of Babesia canis and Babesia vogeli.Ticks Tick Borne Dis. 2017 Oct;8(6):862-865. doi: 10.1016/j.ttbdis.2017.07.002. Epub 2017 Jul 8. Ticks Tick Borne Dis. 2017. PMID: 28739301
-
Tick-borne haemoparasites and Anaplasmataceae in domestic dogs in Zambia.Ticks Tick Borne Dis. 2018 May;9(4):988-995. doi: 10.1016/j.ttbdis.2018.03.025. Epub 2018 Mar 26. Ticks Tick Borne Dis. 2018. PMID: 29622515
-
Molecular investigation of tick-borne pathogens in dogs from Luanda, Angola.Parasit Vectors. 2016 May 10;9(1):252. doi: 10.1186/s13071-016-1536-z. Parasit Vectors. 2016. PMID: 27160839 Free PMC article.
-
An annotated checklist of tick-borne pathogens of dogs in Nigeria.Vet Parasitol Reg Stud Reports. 2019 Jan;15:100255. doi: 10.1016/j.vprsr.2018.12.001. Epub 2018 Dec 3. Vet Parasitol Reg Stud Reports. 2019. PMID: 30929932 Review.
-
A Review on NanoPCR: History, Mechanism and Applications.J Nanosci Nanotechnol. 2018 Dec 1;18(12):8029-8046. doi: 10.1166/jnn.2018.16390. J Nanosci Nanotechnol. 2018. PMID: 30189919 Review.
Cited by
-
Application of Nanomaterials to Enhance Polymerase Chain Reaction.Molecules. 2022 Dec 13;27(24):8854. doi: 10.3390/molecules27248854. Molecules. 2022. PMID: 36557991 Free PMC article. Review.
-
Shine: A novel strategy to extract specific, sensitive and well-conserved biomarkers from massive microbial genomic datasets.BMC Bioinformatics. 2023 Apr 4;24(1):128. doi: 10.1186/s12859-023-05195-2. BMC Bioinformatics. 2023. PMID: 37016282 Free PMC article.
-
Nanotechnology-Enabled PCR with Tunable Energy Dynamics.JACS Au. 2024 Aug 30;4(9):3370-3382. doi: 10.1021/jacsau.4c00570. eCollection 2024 Sep 23. JACS Au. 2024. PMID: 39328766 Free PMC article. Review.
-
The nucleic acid reactions on the nanomaterials surface for biomedicine.J Nanobiotechnology. 2025 Apr 23;23(1):308. doi: 10.1186/s12951-025-03374-2. J Nanobiotechnology. 2025. PMID: 40269855 Free PMC article. Review.
-
Nanomaterials in PCR: exploring light-to-heat conversion mechanisms and microfluidic integration.Microsyst Nanoeng. 2025 Jun 19;11(1):127. doi: 10.1038/s41378-025-00898-3. Microsyst Nanoeng. 2025. PMID: 40533488 Free PMC article. Review.
References
-
- Mullis K.B., Faloona F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. - PubMed
-
- Huber M., Mundlein A., Dornstauder E., Schneeberger C., Tempfer C.B., Mueller M.W., Schmidt W.M. Accessing single nucleotide polymorphisms in genomic DNA by direct multiplex polymerase chain reaction amplification on oligonucleotide microarrays. Anal. Biochem. 2002;303:25–33. doi: 10.1006/abio.2001.5565. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

