Short interspersed nuclear element (SINE)-mediated post-transcriptional effects on human and mouse gene expression: SINE-UP for active duty
- PMID: 32075563
- PMCID: PMC7061979
- DOI: 10.1098/rstb.2019.0344
Short interspersed nuclear element (SINE)-mediated post-transcriptional effects on human and mouse gene expression: SINE-UP for active duty
Abstract
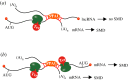
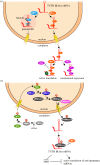
Primate-specific Alu short interspersed nuclear elements (SINEs) and rodent-specific B and ID (B/ID) SINEs are non-autonomous and generally non-coding retrotransposons that have been copied and pasted into the respective genomes so as to constitute what is estimated to be a remarkable 13% and 8% of those genomes. In the context of messenger RNAs (mRNAs), those residing within 3'-untranslated regions (3'UTRs) can influence mRNA export from the nucleus to the cytoplasm, mRNA translation and/or mRNA decay via proteins with which they associate either individually or base-paired in cis or in trans with a partially complementary SINE. Each of these influences impinges on the primary function of mRNA, which is to serve as a template for protein synthesis. This review describes how human cells have used 3'UTR Alu elements to mediate post-transcriptional gene regulation and also describes examples of convergent evolution between human and mouse 3'UTR SINEs. This article is part of a discussion meeting issue 'Crossroads between transposons and gene regulation'.
Keywords: 3′-untranslated regions; double-stranded RNA-binding proteins; mRNA export; mRNA translation, mRNA decay; short interspersed elements.
Conflict of interest statement
I declare I have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
