Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes
- PMID: 32075564
- PMCID: PMC7061989
- DOI: 10.1098/rstb.2019.0347
Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes
Abstract
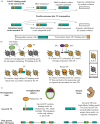
Eukaryotic gene regulation is mediated by cis-regulatory elements, which are embedded within the vast non-coding genomic space and recognized by the transcription factors in a sequence- and context-dependent manner. A large proportion of eukaryotic genomes, including at least half of the human genome, are composed of transposable elements (TEs), which in their ancestral form carried their own cis-regulatory sequences able to exploit the host trans environment to promote TE transcription and facilitate transposition. Although not all present-day TE copies have retained this regulatory function, the preexisting regulatory potential of TEs can provide a rich source of cis-regulatory innovation for the host. Here, we review recent evidence documenting diverse contributions of TE sequences to gene regulation by functioning as enhancers, promoters, silencers and boundary elements. We discuss how TE-derived enhancer sequences can rapidly facilitate changes in existing gene regulatory networks and mediate species- and cell-type-specific regulatory innovations, and we postulate a unique contribution of TEs to species-specific gene expression divergence in pluripotency and early embryogenesis. With advances in genome-wide technologies and analyses, systematic investigation of TEs' cis-regulatory potential is now possible and our understanding of the biological impact of genomic TEs is increasing. This article is part of a discussion meeting issue 'Crossroads between transposons and gene regulation'.
Keywords: enhancers; hourglass model and gene regulation; transcription factor binding; transposons.
Conflict of interest statement
We have no competing interests to declare.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
