How Nanopore Translocation Experiments Can Measure RNA Unfolding
- PMID: 32075749
- PMCID: PMC7136281
- DOI: 10.1016/j.bpj.2020.01.030
How Nanopore Translocation Experiments Can Measure RNA Unfolding
Abstract
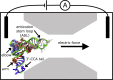
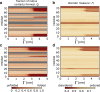
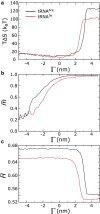
Electrokinetic translocation of biomolecules through solid-state nanopores represents a label-free single-molecule technique that may be used to measure biomolecular structure and dynamics. Recent investigations have attempted to distinguish individual transfer RNA (tRNA) species based on the associated pore translocation times, ion-current noise, and blockage currents. By manufacturing sufficiently smaller pores, each tRNA is required to undergo a deformation to translocate. Accordingly, differences in nanopore translocation times and distributions may be used to infer the mechanical properties of individual tRNA molecules. To bridge our understanding of tRNA structural dynamics and nanopore measurements, we apply molecular dynamics simulations using a simplified "structure-based" energetic model. Calculating the free-energy landscape for distinct tRNA species implicates transient unfolding of the terminal RNA helix during nanopore translocation. This provides a structural and energetic framework for interpreting current experiments, which can aid the design of methods for identifying macromolecules using nanopores.
Copyright © 2020 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Dekker C. Solid-state nanopores. Nat. Nanotechnol. 2007;2:209–215. - PubMed
-
- Venkatesan B.M., Bashir R. Nanopore sensors for nucleic acid analysis. Nat. Nanotechnol. 2011;6:615–624. - PubMed
-
- Miles B.N., Ivanov A.P., Edel J.B. Single molecule sensing with solid-state nanopores: novel materials, methods, and applications. Chem. Soc. Rev. 2013;42:15–28. - PubMed
-
- Spitzberg J.D., Zrehen A., Meller A. Plasmonic-nanopore biosensors for superior single-molecule detection. Adv. Mater. 2019;31:e1900422. - PubMed
-
- Danda G., Drndić M. Two-dimensional nanopores and nanoporous membranes for ion and molecule transport. Curr. Opin. Biotechnol. 2019;55:124–133. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

