The C-terminal domain of feline and bovine SAMHD1 proteins has a crucial role in lentiviral restriction
- PMID: 32075911
- PMCID: PMC7105322
- DOI: 10.1074/jbc.RA120.012767
The C-terminal domain of feline and bovine SAMHD1 proteins has a crucial role in lentiviral restriction
Abstract
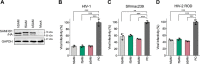
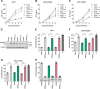
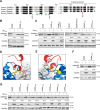
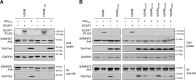
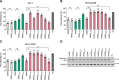
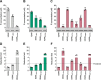
SAM and HD domain-containing protein 1 (SAMHD1) is a host factor that restricts reverse transcription of lentiviruses such as HIV in myeloid cells and resting T cells through its dNTP triphosphohydrolase (dNTPase) activity. Lentiviruses counteract this restriction by expressing the accessory protein Vpx or Vpr, which targets SAMHD1 for proteasomal degradation. SAMHD1 is conserved among mammals, and the feline and bovine SAMHD1 proteins (fSAM and bSAM) restrict lentiviruses by reducing cellular dNTP concentrations. However, the functional regions of fSAM and bSAM that are required for their biological functions are not well-characterized. Here, to establish alternative models to investigate SAMHD1 in vivo, we studied the restriction profile of fSAM and bSAM against different primate lentiviruses. We found that both fSAM and bSAM strongly restrict primate lentiviruses and that Vpx induces the proteasomal degradation of both fSAM and bSAM. Further investigation identified one and five amino acid sites in the C-terminal domain (CTD) of fSAM and bSAM, respectively, that are required for Vpx-mediated degradation. We also found that the CTD of bSAM is directly involved in mediating bSAM's antiviral activity by regulating dNTPase activity, whereas the CTD of fSAM is not. Our results suggest that the CTDs of fSAM and bSAM have important roles in their antiviral functions. These findings advance our understanding of the mechanism of fSAM- and bSAM-mediated viral restriction and might inform strategies for improving HIV animal models.
Keywords: AIDS; C-terminal domain; SAM domain and HD domain-containing protein 1 (SAMHD1); Vpx; bovine SAMHD1; dNTPase; degradation; feline SAMHD1; host-pathogen interaction; human immunodeficiency virus (HIV); lentivirus; triphosphohydrolase; viral replication; viral restriction.
© 2020 Wang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Role of Intracellular Distribution of Feline and Bovine SAMHD1 Proteins in Lentiviral Restriction.Virol Sin. 2021 Oct;36(5):981-996. doi: 10.1007/s12250-021-00351-5. Epub 2021 Mar 22. Virol Sin. 2021. PMID: 33751400 Free PMC article.
-
Interplay of ancestral non-primate lentiviruses with the virus-restricting SAMHD1 proteins of their hosts.J Biol Chem. 2018 Oct 19;293(42):16402-16412. doi: 10.1074/jbc.RA118.004567. Epub 2018 Sep 4. J Biol Chem. 2018. PMID: 30181218 Free PMC article.
-
Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells.Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):2729-2734. doi: 10.1073/pnas.1613635114. Epub 2017 Feb 22. Proc Natl Acad Sci U S A. 2017. PMID: 28228523 Free PMC article.
-
[Research Progress in Viral Protein Vpx induction of Proteasomal Degradation of the Antiviral Factor SAMHD1].Bing Du Xue Bao. 2016 May;32(3):355-60. Bing Du Xue Bao. 2016. PMID: 29963825 Review. Chinese.
-
Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase.Curr Opin Virol. 2012 Dec;2(6):755-63. doi: 10.1016/j.coviro.2012.09.010. Epub 2012 Oct 10. Curr Opin Virol. 2012. PMID: 23062609 Free PMC article. Review.
Cited by
-
SAMHD1 Functions and Human Diseases.Viruses. 2020 Mar 31;12(4):382. doi: 10.3390/v12040382. Viruses. 2020. PMID: 32244340 Free PMC article. Review.
-
Retroviral Restriction Factors and Their Viral Targets: Restriction Strategies and Evolutionary Adaptations.Microorganisms. 2020 Dec 11;8(12):1965. doi: 10.3390/microorganisms8121965. Microorganisms. 2020. PMID: 33322320 Free PMC article. Review.
-
Role of Intracellular Distribution of Feline and Bovine SAMHD1 Proteins in Lentiviral Restriction.Virol Sin. 2021 Oct;36(5):981-996. doi: 10.1007/s12250-021-00351-5. Epub 2021 Mar 22. Virol Sin. 2021. PMID: 33751400 Free PMC article.
-
Comprehensive Investigation on the Interplay between Feline APOBEC3Z3 Proteins and Feline Immunodeficiency Virus Vif Proteins.J Virol. 2021 Jun 10;95(13):e0017821. doi: 10.1128/JVI.00178-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33762419 Free PMC article.
-
Host cell restriction factors of equine infectious anemia virus.Virol Sin. 2023 Aug;38(4):485-496. doi: 10.1016/j.virs.2023.07.001. Epub 2023 Jul 5. Virol Sin. 2023. PMID: 37419416 Free PMC article. Review.
References
-
- Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., and Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 10.1038/nature10623 - DOI - PubMed
-
- Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., and Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 10.1038/ni.2236 - DOI - PMC - PubMed
-
- St. Gelais C., de Silva S., Amie S. M., Coleman C. M., Hoy H., Hollenbaugh J. A., Kim B., and Wu L. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105 10.1186/1742-4690-9-105 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

