A molecular gradient along the longitudinal axis of the human hippocampus informs large-scale behavioral systems
- PMID: 32075960
- PMCID: PMC7031290
- DOI: 10.1038/s41467-020-14518-3
A molecular gradient along the longitudinal axis of the human hippocampus informs large-scale behavioral systems
Abstract
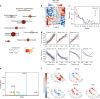
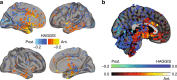
The functional organization of the hippocampus is distributed as a gradient along its longitudinal axis that explains its differential interaction with diverse brain systems. We show that the location of human tissue samples extracted along the longitudinal axis of the adult human hippocampus can be predicted within 2mm using the expression pattern of less than 100 genes. Futhermore, this model generalizes to an external set of tissue samples from prenatal human hippocampi. We examine variation in this specific gene expression pattern across the whole brain, finding a distinct anterioventral-posteriodorsal gradient. We find frontal and anterior temporal regions involved in social and motivational behaviors, and more functionally connected to the anterior hippocampus, to be clearly differentiated from posterior parieto-occipital regions involved in visuospatial cognition and more functionally connected to the posterior hippocampus. These findings place the human hippocampus at the interface of two major brain systems defined by a single molecular gradient.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Andersen, P., Morris, R., Amaral, D., Bliss, T. and O’Keefe, J. The Hippocampus Book (Oxford University Press, 2006) 10.1093/acprof:oso/9780195100273.001.0001/acprof-9780195100273.

