DNA extraction of microbial DNA directly from infected tissue: an optimized protocol for use in nanopore sequencing
- PMID: 32076089
- PMCID: PMC7031281
- DOI: 10.1038/s41598-020-59957-6
DNA extraction of microbial DNA directly from infected tissue: an optimized protocol for use in nanopore sequencing
Abstract
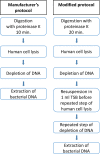
Identification of bacteria causing tissue infections can be comprehensive and, in the cases of non- or slow-growing bacteria, near impossible with conventional methods. Performing shotgun metagenomic sequencing on bacterial DNA extracted directly from the infected tissue may improve time to diagnosis and targeted treatment considerably. However, infected tissue consists mainly of human DNA (hDNA) which hampers bacterial identification. In this proof of concept study, we present a modified version of the Ultra-Deep Microbiome Prep kit for DNA extraction procedure, removing additional human DNA. Tissue biopsies from 3 patients with orthopedic implant-related infections containing varying degrees of Staphylococcus aureus were included. Subsequent DNA shotgun metagenomic sequencing using Oxford Nanopore Technologies' (ONT) MinION platform and ONTs EPI2ME WIMP and ARMA bioinformatic workflows for microbe and antibiotic resistance genes identification, respectively. The modified DNA extraction protocol led to an additional ~10-fold reduction of human DNA while preserving S. aureus DNA. Including the DNA sequencing and bioinformatics analyses, the presented protocol has the potential of identifying the infection-causing pathogen in infected tissue within 7 hours after biopsy. However, due to low number of S. aureus reads, positive identification of antibiotic resistance genes was not possible.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Evaluation of DNA extraction kits for long-read shotgun metagenomics using Oxford Nanopore sequencing for rapid taxonomic and antimicrobial resistance detection.Sci Rep. 2024 Nov 27;14(1):29531. doi: 10.1038/s41598-024-80660-3. Sci Rep. 2024. PMID: 39604411 Free PMC article.
-
Shotgun-metagenomics based prediction of antibiotic resistance and virulence determinants in Staphylococcus aureus from periprosthetic tissue on blood culture bottles.Sci Rep. 2021 Oct 21;11(1):20848. doi: 10.1038/s41598-021-00383-7. Sci Rep. 2021. PMID: 34675288 Free PMC article.
-
Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing.J Microbiol Methods. 2016 Aug;127:141-145. doi: 10.1016/j.mimet.2016.05.022. Epub 2016 May 26. J Microbiol Methods. 2016. PMID: 27237775 Free PMC article.
-
Concise survey of Staphylococcus aureus virulence factors that promote adhesion and damage to peri-implant tissues.Int J Artif Organs. 2011 Sep;34(9):771-80. doi: 10.5301/ijao.5000046. Int J Artif Organs. 2011. PMID: 22094556 Review.
-
Emerging strategies and therapeutic innovations for combating drug resistance in Staphylococcus aureus strains: A comprehensive review.J Basic Microbiol. 2024 May;64(5):e2300579. doi: 10.1002/jobm.202300579. Epub 2024 Feb 2. J Basic Microbiol. 2024. PMID: 38308076 Review.
Cited by
-
Characterization of Fecal Microbiota with Clinical Specimen Using Long-Read and Short-Read Sequencing Platform.Int J Mol Sci. 2020 Sep 26;21(19):7110. doi: 10.3390/ijms21197110. Int J Mol Sci. 2020. PMID: 32993155 Free PMC article.
-
Development of an Amplicon Nanopore Sequencing Strategy for Detection of Mutations Conferring Intermediate Resistance to Vancomycin in Staphylococcus aureus Strains.Microbiol Spectr. 2023 Feb 14;11(1):e0272822. doi: 10.1128/spectrum.02728-22. Epub 2023 Jan 23. Microbiol Spectr. 2023. PMID: 36688645 Free PMC article.
-
Rapid metagenomic sequencing for diagnosis and antimicrobial sensitivity prediction of canine bacterial infections.Microb Genom. 2023 Jul;9(7):mgen001066. doi: 10.1099/mgen.0.001066. Microb Genom. 2023. PMID: 37471128 Free PMC article.
-
An Efficient and Easy- to- Use Method for Extraction of H. pylori DNA from Archival Formalin-Fixed and Paraffin-Embedded Gastric Tissues.Iran J Pathol. 2023;18(4):410-414. doi: 10.30699/IJP.2023.562113.2974. Epub 2023 Oct 15. Iran J Pathol. 2023. PMID: 38024551 Free PMC article.
-
Effect of bacterial DNA enrichment on detection and quantification of bacteria in an infected tissue model by metagenomic next-generation sequencing.ISME Commun. 2022 Dec 26;2(1):122. doi: 10.1038/s43705-022-00208-2. ISME Commun. 2022. PMID: 37938717 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous