ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity
- PMID: 32076273
- PMCID: PMC7060130
- DOI: 10.1038/s41586-020-2015-4
ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity
Abstract
Group 2 innate lymphoid cells (ILC2s) regulate inflammation and immunity in mammalian tissues1,2. Although ILC2s are found in cancers of these tissues3, their roles in cancer immunity and immunotherapy are unclear. Here we show that ILC2s infiltrate pancreatic ductal adenocarcinomas (PDACs) to activate tissue-specific tumour immunity. Interleukin-33 (IL33) activates tumour ILC2s (TILC2s) and CD8+ T cells in orthotopic pancreatic tumours but not heterotopic skin tumours in mice to restrict pancreas-specific tumour growth. Resting and activated TILC2s express the inhibitory checkpoint receptor PD-1. Antibody-mediated PD-1 blockade relieves ILC2 cell-intrinsic PD-1 inhibition to expand TILC2s, augment anti-tumour immunity, and enhance tumour control, identifying activated TILC2s as targets of anti-PD-1 immunotherapy. Finally, both PD-1+ TILC2s and PD-1+ T cells are present in most human PDACs. Our results identify ILC2s as anti-cancer immune cells for PDAC immunotherapy. More broadly, ILC2s emerge as tissue-specific enhancers of cancer immunity that amplify the efficacy of anti-PD-1 immunotherapy. As ILC2s and T cells co-exist in human cancers and share stimulatory and inhibitory pathways, immunotherapeutic strategies to collectively target anti-cancer ILC2s and T cells may be broadly applicable.
Conflict of interest statement
Figures




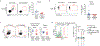







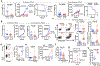

Comment in
-
IL33 Stimulates Innate Lymphoid Cells to Recruit CD8+ T Cells to PDACs.Cancer Discov. 2020 Apr;10(4):489. doi: 10.1158/2159-8290.CD-RW2020-032. Epub 2020 Feb 28. Cancer Discov. 2020. PMID: 32111599
-
Immuno-Oncology beyond TILs: Unleashing TILCs.Cancer Cell. 2020 Apr 13;37(4):428-430. doi: 10.1016/j.ccell.2020.03.021. Cancer Cell. 2020. PMID: 32289267
-
Enabling anti-tumor immunity by unleashing ILC2.Cell Res. 2020 Jun;30(6):461-462. doi: 10.1038/s41422-020-0330-9. Cell Res. 2020. PMID: 32367044 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

