Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity
- PMID: 32076642
- PMCID: PMC7002122
- DOI: 10.1126/sciadv.aay0187
Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity
Abstract
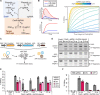
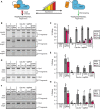
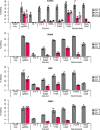
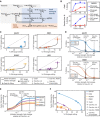
The limited target specificity of CRISPR-Cas nucleases poses a challenge with respect to their application in research and therapy. Here, we present a simple and original strategy to enhance the specificity of CRISPR-Cas9 genome editing by coupling Cas9 to artificial inhibitory domains. Applying a combination of mathematical modeling and experiments, we first determined how CRISPR-Cas9 activity profiles relate to Cas9 specificity. We then used artificially weakened anti-CRISPR (Acr) proteins either coexpressed with or directly fused to Cas9 to fine-tune its activity toward selected levels, thereby achieving an effective kinetic insulation of ON- and OFF-target editing events. We demonstrate highly specific genome editing in mammalian cells using diverse single-guide RNAs prone to potent OFF-targeting. Last, we show that our strategy is compatible with different modes of delivery, including transient transfection and adeno-associated viral vectors. Together, we provide a highly versatile approach to reduce CRISPR-Cas OFF-target effects via kinetic insulation.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures
References
-
- Amrani N., Gao X. D., Liu P., Edraki A., Mir A., Ibraheim R., Gupta A., Sasaki K. E., Wu T., Donohoue P. D., Settle A. H., Lied A. M., McGovern K., Fuller C. K., Cameron P., Fazzio T. G., Zhu L. J., Wolfe S. A., Sontheimer E. J., NmeCas9 is an intrinsically high-fidelity genome-editing platform. Genome Biol. 19, 214 (2018). - PMC - PubMed
-
- Kim D., Kim J., Hur J. K., Been K. W., Yoon S.-H., Kim J.-S., Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

