Decoding a highly mixed Kazakh genome
- PMID: 32076829
- PMCID: PMC7170836
- DOI: 10.1007/s00439-020-02132-8
Decoding a highly mixed Kazakh genome
Abstract
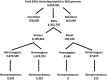
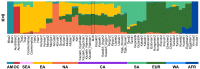
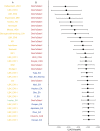
We provide a Kazakh whole genome sequence (MJS) and analyses with the largest comparative Kazakh genomic data available to date. We found 102,240 novel SNVs and a high level of heterozygosity. ADMIXTURE analysis confirmed a significant proportion of variations in this individual coming from all continents except Africa and Oceania. A principal component analysis showed neighboring Kalmyk, Uzbek, and Kyrgyz populations to have the strongest resemblance to the MJS genome which reflects fairly recent Kazakh history. MJS's mitochondrial haplogroup, J1c2, probably represents an early European and Near Eastern influence to Central Asia. This was also supported by the heterozygous SNPs associated with European phenotypic features and strikingly similar Kazakh ancestral composition inferred by ADMIXTURE. Admixture (f3) analysis showed that MJS's genomic signature is best described as a cross between the Neolithic East Asian (Devil's Gate1) and the Bronze Age European (Halberstadt_LBA1) components rather than a contemporary admixture.
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures

References
-
- Akerov TA. On the origin of the Naiman. J Siberian Fed Univ. 2016;9(9):2071–2081. doi: 10.17516/1997-1370-2016-9-9-2071-2081. - DOI
-
- Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, et al. Population genomics of bronze age Eurasia. Nature. 2015;522:167. - PubMed
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

