Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration
- PMID: 32078387
- PMCID: PMC7241614
- DOI: 10.1161/CIRCULATIONAHA.119.043067
Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration
Abstract
Background: The adult mammalian heart has limited regenerative capacity, mostly attributable to postnatal cardiomyocyte cell cycle arrest. In the last 2 decades, numerous studies have explored cardiomyocyte cell cycle regulatory mechanisms to enhance myocardial regeneration after myocardial infarction. Pkm2 (Pyruvate kinase muscle isoenzyme 2) is an isoenzyme of the glycolytic enzyme pyruvate kinase. The role of Pkm2 in cardiomyocyte proliferation, heart development, and cardiac regeneration is unknown.
Methods: We investigated the effect of Pkm2 in cardiomyocytes through models of loss (cardiomyocyte-specific Pkm2 deletion during cardiac development) or gain using cardiomyocyte-specific Pkm2 modified mRNA to evaluate Pkm2 function and regenerative affects after acute or chronic myocardial infarction in mice.
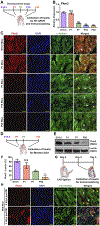
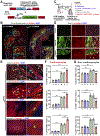
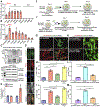
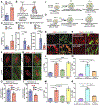
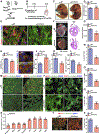
Results: Here, we identify Pkm2 as an important regulator of the cardiomyocyte cell cycle. We show that Pkm2 is expressed in cardiomyocytes during development and immediately after birth but not during adulthood. Loss of function studies show that cardiomyocyte-specific Pkm2 deletion during cardiac development resulted in significantly reduced cardiomyocyte cell cycle, cardiomyocyte numbers, and myocardial size. In addition, using cardiomyocyte-specific Pkm2 modified RNA, our novel cardiomyocyte-targeted strategy, after acute or chronic myocardial infarction, resulted in increased cardiomyocyte cell division, enhanced cardiac function, and improved long-term survival. We mechanistically show that Pkm2 regulates the cardiomyocyte cell cycle and reduces oxidative stress damage through anabolic pathways and β-catenin.
Conclusions: We demonstrate that Pkm2 is an important intrinsic regulator of the cardiomyocyte cell cycle and oxidative stress, and highlight its therapeutic potential using cardiomyocyte-specific Pkm2 modified RNA as a gene delivery platform.
Keywords: anabolism; cardiomyocytes; cell proliferation; regeneration.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

