Comprehensive assessment of computational algorithms in predicting cancer driver mutations
- PMID: 32079540
- PMCID: PMC7033911
- DOI: 10.1186/s13059-020-01954-z
Comprehensive assessment of computational algorithms in predicting cancer driver mutations
Abstract
Background: The initiation and subsequent evolution of cancer are largely driven by a relatively small number of somatic mutations with critical functional impacts, so-called driver mutations. Identifying driver mutations in a patient's tumor cells is a central task in the era of precision cancer medicine. Over the decade, many computational algorithms have been developed to predict the effects of missense single-nucleotide variants, and they are frequently employed to prioritize mutation candidates. These algorithms employ diverse molecular features to build predictive models, and while some algorithms are cancer-specific, others are not. However, the relative performance of these algorithms has not been rigorously assessed.
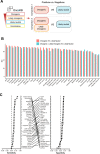
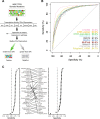
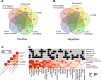
Results: We construct five complementary benchmark datasets: mutation clustering patterns in the protein 3D structures, literature annotation based on OncoKB, TP53 mutations based on their effects on target-gene transactivation, effects of cancer mutations on tumor formation in xenograft experiments, and functional annotation based on in vitro cell viability assays we developed including a new dataset of ~ 200 mutations. We evaluate the performance of 33 algorithms and found that CHASM, CTAT-cancer, DEOGEN2, and PrimateAI show consistently better performance than the other algorithms. Moreover, cancer-specific algorithms show much better performance than those designed for a general purpose.
Conclusions: Our study is a comprehensive assessment of the performance of different algorithms in predicting cancer driver mutations and provides deep insights into the best practice of computationally prioritizing cancer mutation candidates for end-users and for the future development of new algorithms.
Keywords: 3D clustering; Cell viability assay; Driver mutations; Passenger mutations; TP53 mutations; The Cancer Genome Atlas; Tumor transformation.
Conflict of interest statement
G.B.M. is on the Scientific Advisory Board for AstraZeneca, ImmunoMet, Nuevolution, and Precision Medicine. H.L. is a shareholder and on the Scientific Advisory Board for Precision Scientific Ltd. The other authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

