Regulation of intrinsic polarity establishment by a differentiation-type MAPK pathway in S. cerevisiae
- PMID: 32079658
- PMCID: PMC7174846
- DOI: 10.1242/jcs.241513
Regulation of intrinsic polarity establishment by a differentiation-type MAPK pathway in S. cerevisiae
Abstract
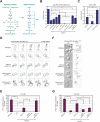
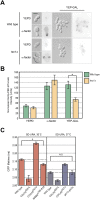
All cells establish and maintain an axis of polarity that is critical for cell shape and progression through the cell cycle. A well-studied example of polarity establishment is bud emergence in the yeast Saccharomyces cerevisiae, which is controlled by the Rho GTPase Cdc42p. The prevailing view of bud emergence does not account for regulation by extrinsic cues. Here, we show that the filamentous growth mitogen activated protein kinase (fMAPK) pathway regulates bud emergence under nutrient-limiting conditions. The fMAPK pathway regulated the expression of polarity targets including the gene encoding a direct effector of Cdc42p, Gic2p. The fMAPK pathway also stimulated GTP-Cdc42p levels, which is a critical determinant of polarity establishment. The fMAPK pathway activity was spatially restricted to bud sites and active during the period of the cell cycle leading up to bud emergence. Time-lapse fluorescence microscopy showed that the fMAPK pathway stimulated the rate of bud emergence during filamentous growth. Unregulated activation of the fMAPK pathway induced multiple rounds of symmetry breaking inside the growing bud. Collectively, our findings identify a new regulatory aspect of bud emergence that sensitizes this essential cellular process to external cues.
Keywords: Bud emergence; Cdc42p; MAPK; Polarity establishment; Pseudohyphal growth; Symmetry breaking.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures




References
-
- Adhikari H., Vadaie N., Chow J., Caccamise L. M., Chavel C. A., Li B., Bowitch A., Stefan C. J. and Cullen P. J. (2015). Role of the unfolded protein response in regulating the mucin-dependent filamentous-growth mitogen-activated protein kinase pathway. Mol. Cell. Biol. 35, 1414-1432. 10.1128/MCB.01501-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

