Immune cell constitution in the tumor microenvironment predicts the outcome in diffuse large B-cell lymphoma
- PMID: 32079690
- PMCID: PMC7927991
- DOI: 10.3324/haematol.2019.243626
Immune cell constitution in the tumor microenvironment predicts the outcome in diffuse large B-cell lymphoma
Abstract
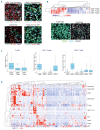
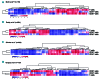
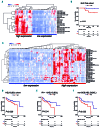
The tumor microenvironment (TME) and limited immune surveillance play important roles in lymphoma pathogenesis. Here we aimed to characterize immunological profiles of diffuse large B-cell lymphoma (DLBCL) and predict the outcome in response to immunochemotherapy. We profiled the expression of 730 immune-related genes in tumor tissues of 81 patients with DLBCL utilizing the Nanostring platform, and used multiplex immunohistochemistry to characterize T-cell phenotypes, including cytotoxic T cells (CD8, Granzyme B, OX40, Ki67), T-cell immune checkpoint (CD3, CD4, CD8, PD1, TIM3, LAG3), as well as regulatory T-cells and Th1 effector cells (CD3, CD4, FOXP3, TBET) in 188 patients. We observed a high degree of heterogeneity at the transcriptome level. Correlation matrix analysis identified gene expression signatures with highly correlating genes, the main cluster containing genes for cytolytic factors, immune checkpoint molecules, T cells and macrophages, together named a TME immune cell signature. Immunophenotyping of the distinct cell subsets revealed that a high proportion of immune checkpoint positive T cells translated to unfavorable survival. Together, our results demonstrate that the immunological profile of DLBCL TME is heterogeneous and clinically meaningful. This highlights the potential impact of T-cell immune checkpoint in regulating survival and resistance to immunochemotherapy. (Registered at clinicaltrials.gov identifiers: NCT01502982 and NCT01325194.)
Figures



References
-
- Coiffier B, Lepage E, Briere J, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(4):235-242. - PubMed
-
- Pfreundschuh M, Trumper L, Osterborg A, et al. CHOP-like chemotherapy plus rituximab versus CHOP-like chemotherapy alone in young patients with good-prognosis diffuse large-B-cell lymphoma: a randomised controlled trial by the MabThera International Trial (MInT) Group. Lancet Oncol. 2006;7(5):379-391. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511. - PubMed
-
- Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-Bcell lymphoma. N Engl J Med. 2002;346(25):1937-1947. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials

