Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR
- PMID: 32079768
- PMCID: PMC7171558
- DOI: 10.1126/science.aay9813
Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR
Abstract
Biased agonists of G protein-coupled receptors (GPCRs) preferentially activate a subset of downstream signaling pathways. In this work, we present crystal structures of angiotensin II type 1 receptor (AT1R) (2.7 to 2.9 angstroms) bound to three ligands with divergent bias profiles: the balanced endogenous agonist angiotensin II (AngII) and two strongly β-arrestin-biased analogs. Compared with other ligands, AngII promotes more-substantial rearrangements not only at the bottom of the ligand-binding pocket but also in a key polar network in the receptor core, which forms a sodium-binding site in most GPCRs. Divergences from the family consensus in this region, which appears to act as a biased signaling switch, may predispose the AT1R and certain other GPCRs (such as chemokine receptors) to adopt conformations that are capable of activating β-arrestin but not heterotrimeric Gq protein signaling.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
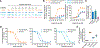
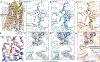
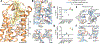

References
-
- Lefkowitz RJ, A brief history of G-protein coupled receptors (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 52, 6366–6378 (2013). - PubMed
-
- Holloway AC et al. , Side-chain substitutions within angiotensin II reveal different requirements for signaling, internalization, and phosphorylation of type 1A angiotensin receptors. Mol. Pharmacol. 61, 768–777 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

