A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield
- PMID: 32080171
- PMCID: PMC7033126
- DOI: 10.1038/s41467-020-14746-7
A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield
Abstract
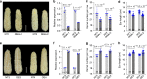
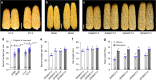
Increasing grain yield of maize (Zea mays L.) is required to meet the rapidly expanding demands for maize-derived food, feed, and fuel. Breeders have enhanced grain productivity of maize hybrids by pyramiding desirable characteristics for larger ears. However, loci selected for improving grain productivity remain largely unclear. Here, we show that a serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 (KNR6) determines pistillate floret number and ear length. Overexpression of KNR6 or introgression of alleles lacking the insertions of two transposable elements in the regulatory region of KNR6 can significantly enhance grain yield. Further in vitro evidences indicate that KNR6 can interact with an Arf GTPase-activating protein (AGAP) and its phosphorylation by KNR6 may affect ear length and kernel number. This finding provides knowledge basis to enhance maize hybrids grain yield.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- McSteen P, Hake S. Barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development. 2001;128:2881–2891. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

