Decimated little brown bats show potential for adaptive change
- PMID: 32080246
- PMCID: PMC7033193
- DOI: 10.1038/s41598-020-59797-4
Decimated little brown bats show potential for adaptive change
Erratum in
-
Author Correction: Decimated little brown bats show potential for adaptive change.Sci Rep. 2020 Mar 24;10(1):5674. doi: 10.1038/s41598-020-62444-7. Sci Rep. 2020. PMID: 32205852 Free PMC article.
Abstract
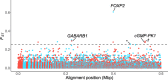
The degree to which species can rapidly adapt is key to survival in the face of climatic and other anthropogenic changes. For little brown bats (Myotis lucifugus), whose populations have experienced declines of over 90% because of the introduced fungal pathogen that causes white-nose syndrome (WNS), survival of the species may ultimately depend upon its capacity for adaptive change. Here, we present evidence of selectively driven change (adaptation), despite dramatic nonadaptive genomic shifts (genetic drift) associated with population declines. We compared the genetic makeups of wild survivors versus non-survivors of WNS, and found significant shifts in allele frequencies of genes associated with regulating arousal from hibernation (GABARB1), breakdown of fats (cGMP-PK1), and vocalizations (FOXP2). Changes at these genes are suggestive of evolutionary adaptation, given that WNS causes bats to arouse with unusual frequency from hibernation, contributing to premature depletion of fat reserves. However, whether these putatively adaptive shifts in allele frequencies translate into sufficient increases in survival for the species to rebound in the face of WNS is unknown.
Conflict of interest statement
The authors declare no competing interests.
Figures

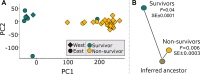
References
-
- Dobson AP, Bradshaw AD, Baker AJM. Hopes for the future: restoration ecology and conservation biology. Sci. 1997;277:515–522. doi: 10.1126/science.277.5325.515. - DOI
-
- Cunningham AA, Daszak P, Rodriguez JP. Pathogen pollution: defining a parasitological threat to biodiversity conservation. J. Parasitology. 2003;89:S78–S83.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

