Chromosome 19 microRNA cluster enhances cell reprogramming by inhibiting epithelial-to-mesenchymal transition
- PMID: 32080251
- PMCID: PMC7033247
- DOI: 10.1038/s41598-020-59812-8
Chromosome 19 microRNA cluster enhances cell reprogramming by inhibiting epithelial-to-mesenchymal transition
Abstract
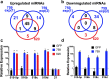
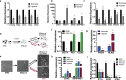
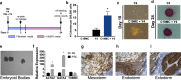
During implantation, cytotrophoblasts undergo epithelial-to-mesenchymal transition (EMT) as they differentiate into invasive extravillous trophoblasts (EVTs). The primate-specific microRNA cluster on chromosome 19 (C19MC) is exclusively expressed in the placenta, embryonic stem cells and certain cancers however, its role in EMT gene regulation is unknown. In situ hybridization for miR-517a/c, a C19MC cistron microRNA, in first trimester human placentas displayed strong expression in villous trophoblasts and a gradual decrease from proximal to distal cell columns as cytotrophoblasts differentiate into invasive EVTs. To investigate the role of C19MC in the regulation of EMT genes, we employed the CRISPR/dCas9 Synergistic Activation Mediator (SAM) system, which induced robust transcriptional activation of the entire C19MC cistron and resulted in suppression of EMT associated genes. Exposure of human iPSCs to hypoxia or differentiation of iPSCs into either cytotrophoblast-stem-like cells or EVT-like cells under hypoxia reduced C19MC expression and increased EMT genes. Furthermore, transcriptional activation of the C19MC cistron induced the expression of OCT4 and FGF4 and accelerated cellular reprogramming. This study establishes the CRISPR/dCas9 SAM as a powerful tool that enables activation of the entire C19MC cistron and uncovers its novel role in suppressing EMT genes critical for maintaining the epithelial cytotrophoblasts stem cell phenotype.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Wang, Y. & Zhao, S. In Vascular Biology of the Placenta Integrated Systems Physiology: from Molecules to Function to Disease (2010).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

