Comparative in-silico proteomic analysis discerns potential granuloma proteins of Yersinia pseudotuberculosis
- PMID: 32080254
- PMCID: PMC7033130
- DOI: 10.1038/s41598-020-59924-1
Comparative in-silico proteomic analysis discerns potential granuloma proteins of Yersinia pseudotuberculosis
Abstract
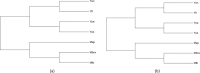
Yersinia pseudotuberculosis is one of the three pathogenic species of the genus Yersinia. Most studies regarding pathogenesis of Y. pseudotuberculosis are based on the proteins related to Type III secretion system, which is a well-known primary virulence factor in pathogenic Gram-negative bacteria, including Y. pseudotuberculosis. Information related to the factors involved in Y. pseudotuberculosis granuloma formation is scarce. In the present study we have used a computational approach to identify proteins that might be potentially involved in formation of Y. pseudotuberculosis granuloma. A comparative proteome analysis and conserved orthologous protein identification was performed between two different genera of bacteria - Mycobacterium and Yersinia, their only common pathogenic trait being ability to form necrotizing granuloma. Comprehensive analysis of orthologous proteins was performed in proteomes of seven bacterial species. This included M. tuberculosis, M. bovis and M. avium paratuberculosis - the known granuloma forming Mycobacterium species, Y. pestis and Y. frederiksenii - the non-granuloma forming Yersinia species and, Y. enterocolitica - that forms micro-granuloma and, Y. pseudotuberculosis - a prominent granuloma forming Yersinia species. In silico proteome analysis indicated that seven proteins (UniProt id A0A0U1QT64, A0A0U1QTE0, A0A0U1QWK3, A0A0U1R1R0, A0A0U1R1Z2, A0A0U1R2S7, A7FMD4) might play some role in Y. pseudotuberculosis granuloma. Validation of the probable involvement of the seven proposed Y. pseudotuberculosis granuloma proteins was done using transcriptome data analysis and, by mapping on a composite protein-protein interaction map of experimentally proved M. tuberculosis granuloma proteins (RD1 locus proteins, ESAT-6 secretion system proteins and intra-macrophage secreted proteins). Though, additional experiments involving knocking out of each of these seven proteins are required to confirm their role in Y. pseudotuberculosis granuloma our study can serve as a basis for further studies on Y. pseudotuberculosis granuloma.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

