UHRF1-repressed 5'-hydroxymethylcytosine is essential for the male meiotic prophase I
- PMID: 32081844
- PMCID: PMC7035279
- DOI: 10.1038/s41419-020-2333-3
UHRF1-repressed 5'-hydroxymethylcytosine is essential for the male meiotic prophase I
Abstract
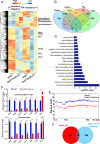
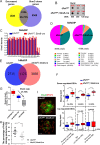
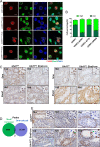
5'-hydroxymethylcytosine (5hmC), an important 5'-cytosine modification, is altered highly in order in male meiotic prophase. However, the regulatory mechanism of this dynamic change and the function of 5hmC in meiosis remain largely unknown. Using a knockout mouse model, we showed that UHRF1 regulated male meiosis. UHRF1 deficiency led to failure of meiosis and male infertility. Mechanistically, the deficiency of UHRF1 altered significantly the meiotic gene profile of spermatocytes. Uhrf1 knockout induced an increase of the global 5hmC level. The enrichment of hyper-5hmC at transcriptional start sites (TSSs) was highly associated with gene downregulation. In addition, the elevated level of the TET1 enzyme might have contributed to the higher 5hmC level in the Uhrf1 knockout spermatocytes. Finally, we reported Uhrf1, a key gene in male meiosis, repressed hyper-5hmC by downregulating TET1. Furthermore, UHRF1 facilitated RNA polymerase II (RNA-pol2) loading to promote gene transcription. Thus our study demonstrated a potential regulatory mechanism of 5hmC dynamic change and its involvement in epigenetic regulation in male meiosis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

