MEN1 deficiency leads to neuroendocrine differentiation of lung cancer and disrupts the DNA damage response
- PMID: 32081882
- PMCID: PMC7035285
- DOI: 10.1038/s41467-020-14614-4
MEN1 deficiency leads to neuroendocrine differentiation of lung cancer and disrupts the DNA damage response
Abstract
The MEN1 gene, a tumor suppressor gene that encodes the protein menin, is mutated at high frequencies in neuroendocrine (NE) tumors; however, the biological importance of this gene in NE-type lung cancer in vivo remains unclear. Here, we established an ATII-specific KrasG12D/+/Men1-/- driven genetically engineered mouse model and show that deficiency of menin results in the accumulation of DNA damage and antagonizes oncogenic Kras-induced senescence and the epithelial-to-mesenchymal transition during lung tumorigenesis. The loss of menin expression in certain human primary lung cancers correlates with elevated NE profiles and reduced overall survival.
Conflict of interest statement
The authors declare no competing interests.
Figures
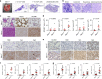


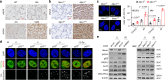
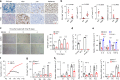
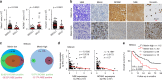

References
-
- II W, AF G, JD M. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001;28:3–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

